IN A NUTSHELL
BastionHub: a universal platform for integrating and analyzing substrates secreted by Gram-negative bacteria
Gram-negative bacteria utilize secretion systems to export substrates into their surrounding environment or directly into neighboring cells. These substrates are proteins that function to promote bacterial survival: by facilitating nutrient collection, disabling competitor species or, for pathogens, to disable host defenses. Following a rapid development of computational techniques, a growing number of substrates have been discovered and subsequently validated by wet lab experiments. To date, several online databases have been developed to catalogue these substrates but they have limited user options for in-depth analysis, and typically focus on a single type of secreted substrate.
In this work, we therefore developed a universal platform, BastionHub, that incorporates extensive functional modules to facilitate substrate analysis and integrates the five major Gram-negative secreted substrate types (i.e. from types I-IV and VI secretion systems). To our knowledge, BastionHub is not only the most comprehensive online database available, it is also the first to incorporate substrates secreted by type I or type II secretion systems. By providing the most up-to-date details of secreted substrates and state-of-the-art prediction and visualized relationship analysis tools, BastionHub will be an important platform that can assist biologists in uncovering novel substrates and formulating new hypotheses.
SUBSTRATE ANALYSIS
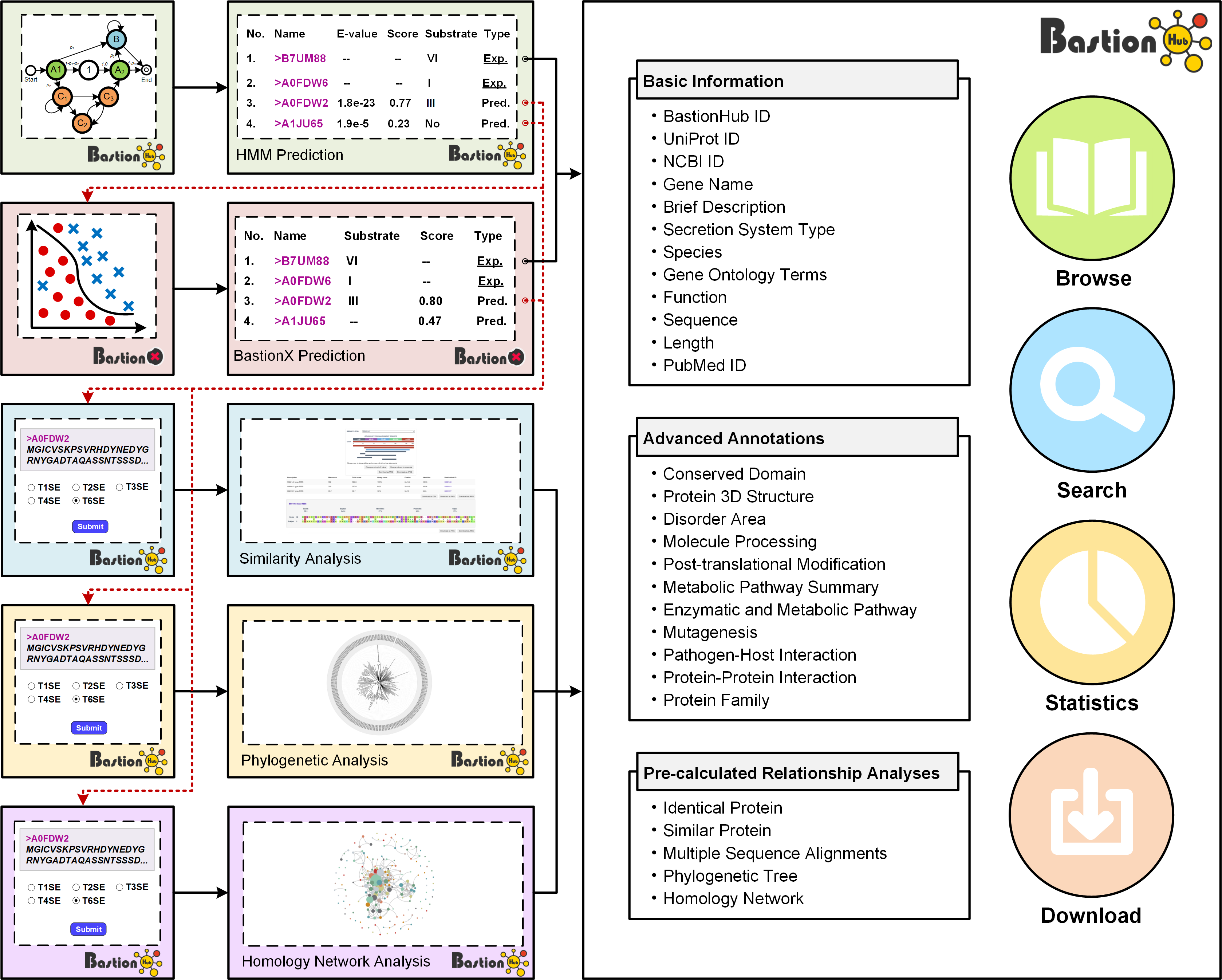
BastionHub incorporates a comprehensive list of manually cuated dataset of type I, II, III, IV, VI secreted substrates, and provides multiple modules for users to investigate them, including browse, search, statistics, download and detailed pages.
1.1 Data Preparation
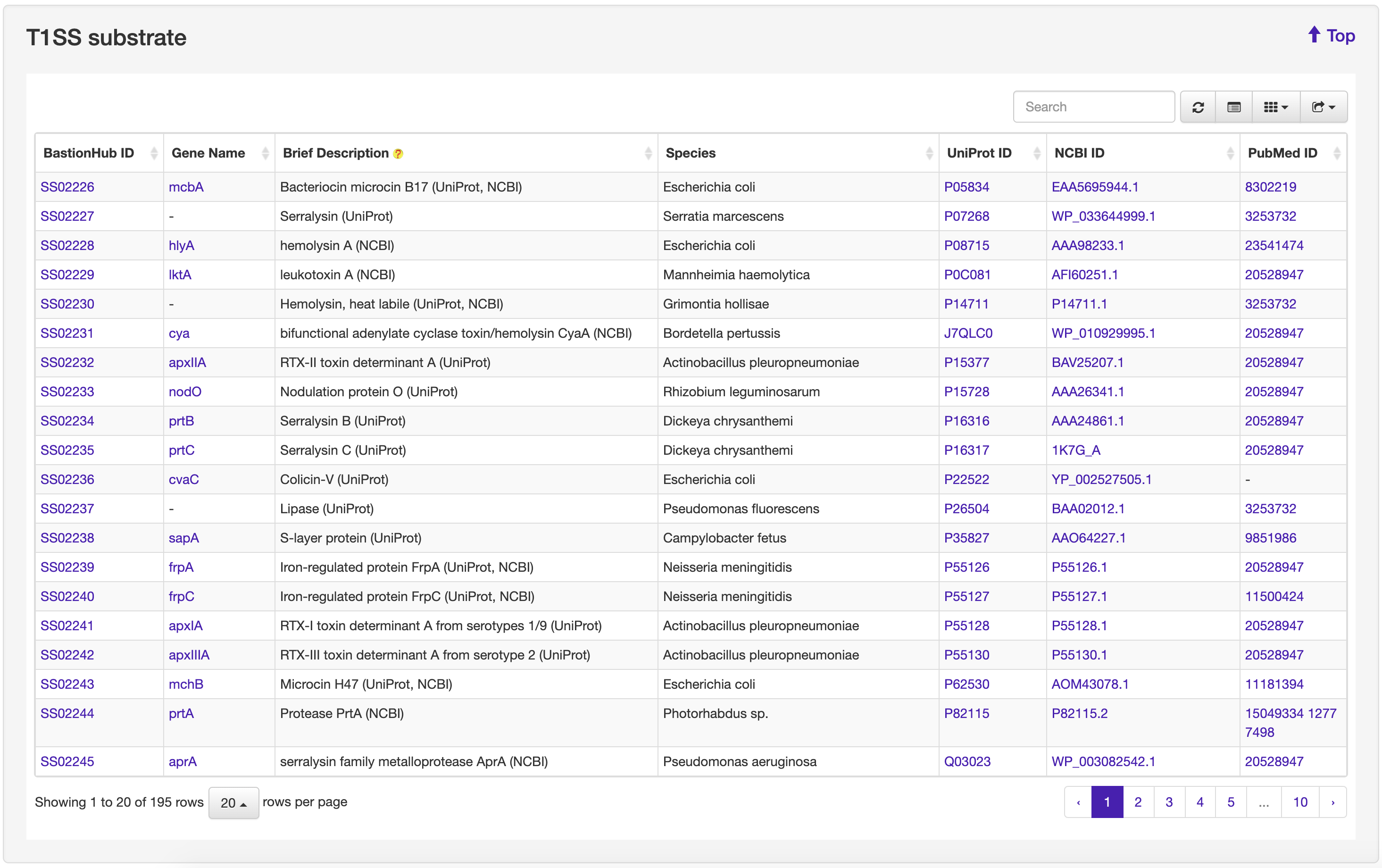
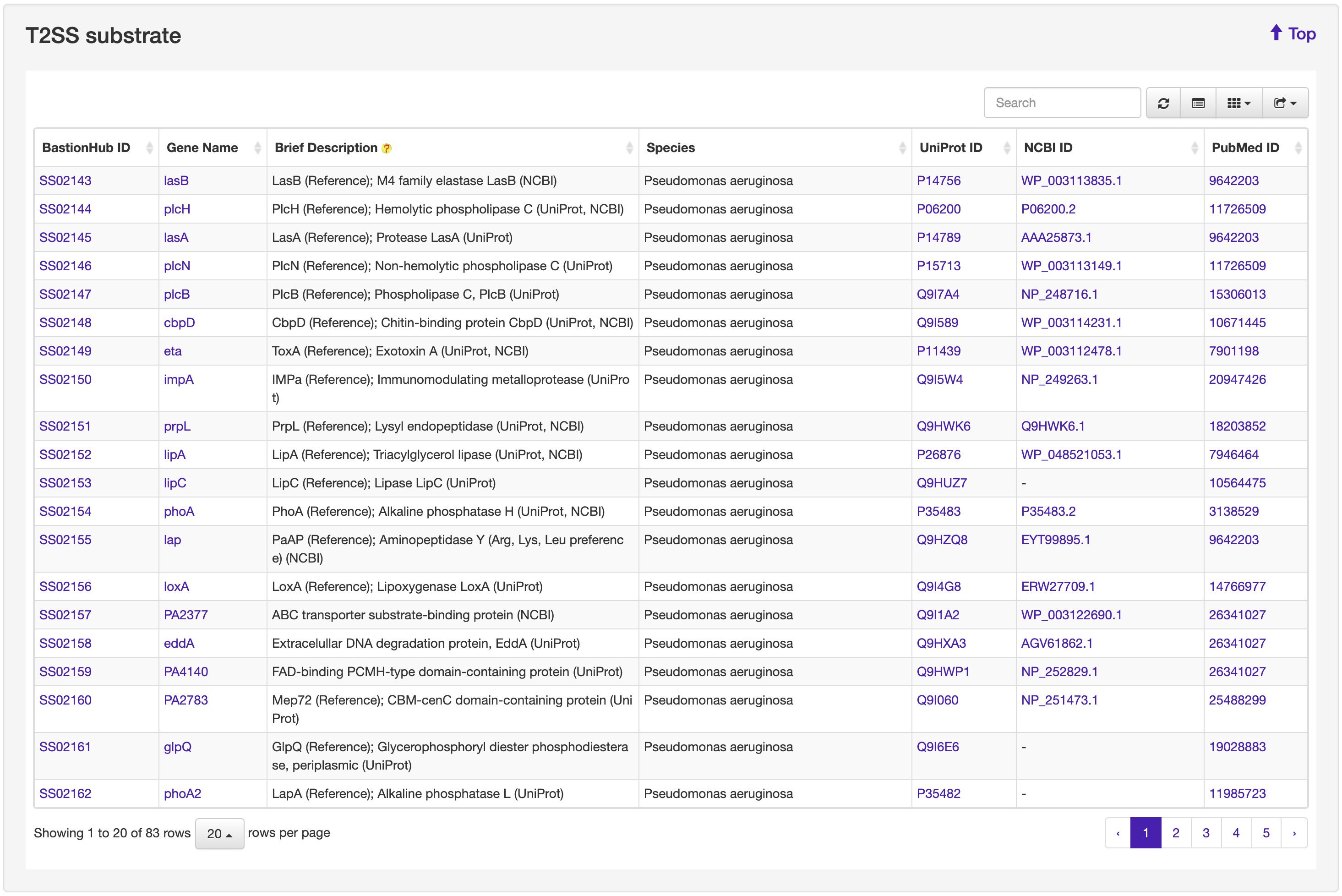
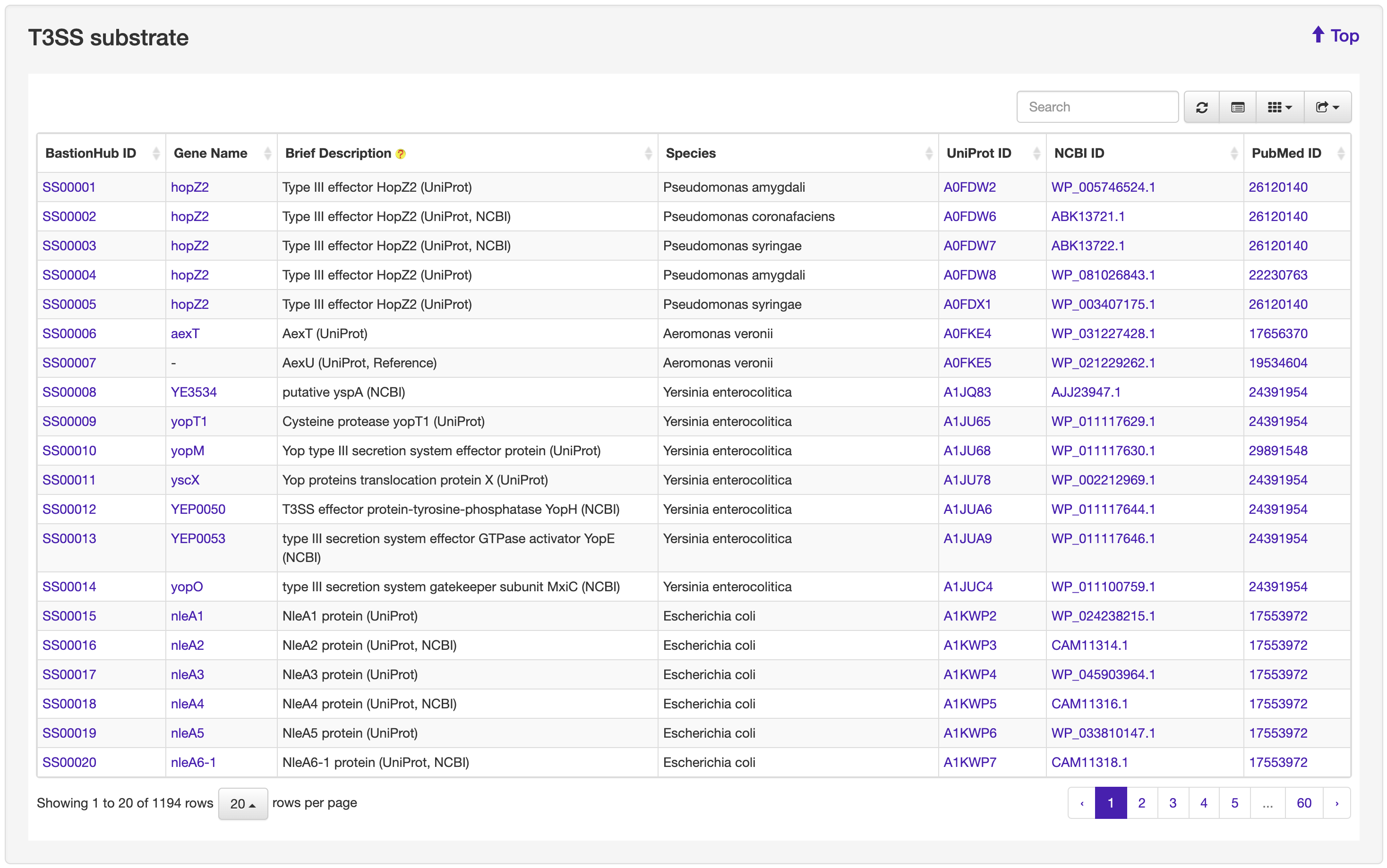
We systematically reviewed existing literature about T1SEs or T2SEs, which was made particularly difficult because there are no uniform names for these secreted substrates. We identified more than 5000 unique references and, after examining each text, we obtained 195 T1SE across 63 species and 83 T2SE across 13 species. From available web resources listing T3SEs, T4SEs, and T6SEs, we extracted details for each substrate to obtain a preliminary dataset. For any entry not annotated with both a UniProt ID and NCBI ID, we used BLAST to identify an identical sequence from the same species to obtain the missing ID code if available. After manually inspecting each individual annotation, we removed obvious errors (e.g. those annotated as “membrane” proteins or “secretion chaperone” proteins). We then annotated the remaining entries with their associated PubMed reference ID where available. Similar to that for T1SE and T2SE, we conducted an exhaustive literature search and retrieved the most recent experimentally validated substrates, including substrates that had previously been overlooked. Accordingly, we obtained 1194 T3SEs across 72 species, 713 T4SEs across 15 species, and 181 T6SEs across 66 species. Altogether, we obtained 2366 substrates secreted by the five secretion systems across 171 species. These substrates were then incorporated into BastionHub, and their annotations can be found on their dedicated ‘Detailed information’ page.
1.2 Browse
The Browse page of BastionHub presents lists of the type I, II, III, IV, VI secreted substrates, which easy-to-use functions to do quick sort, search and download functions.
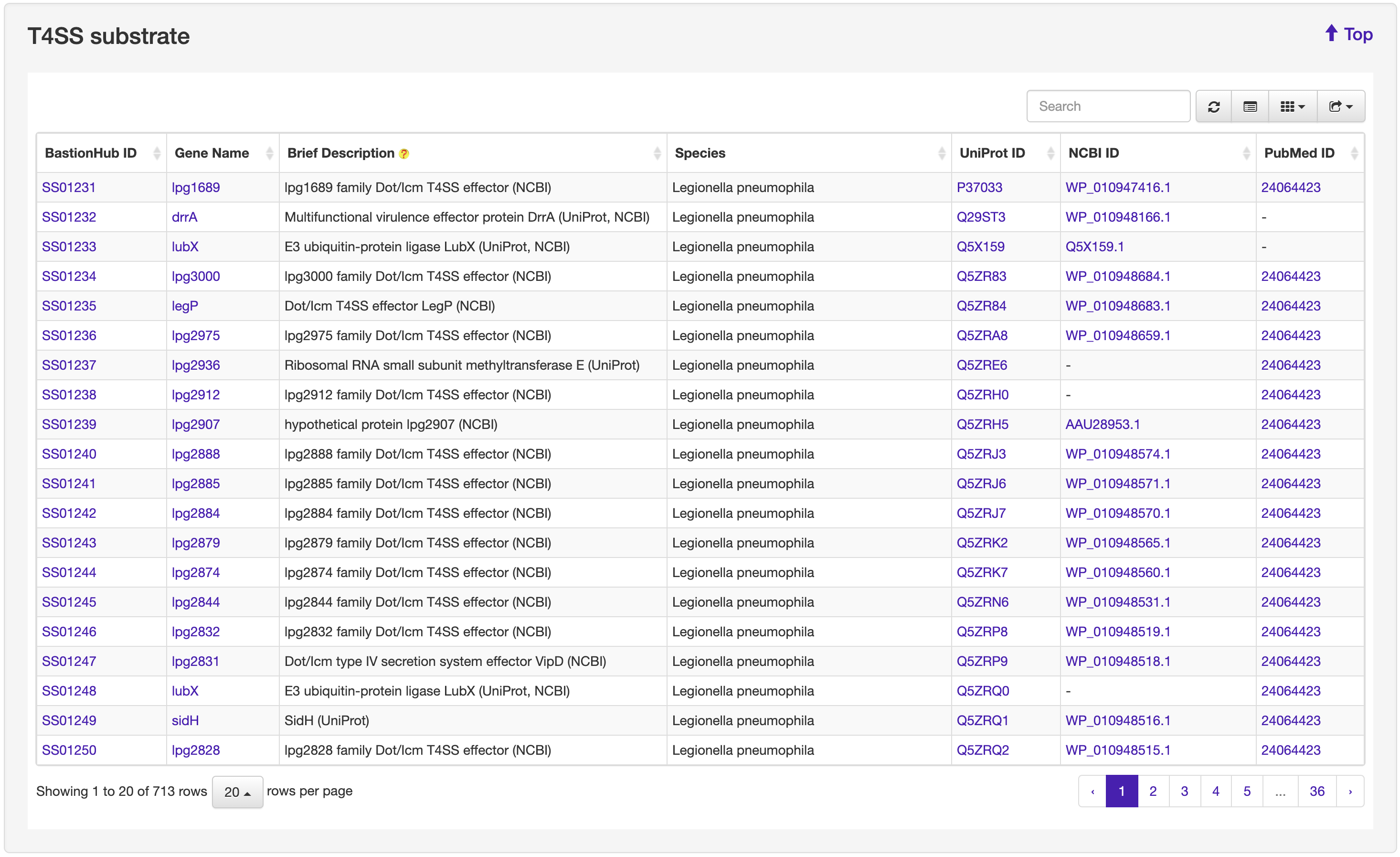
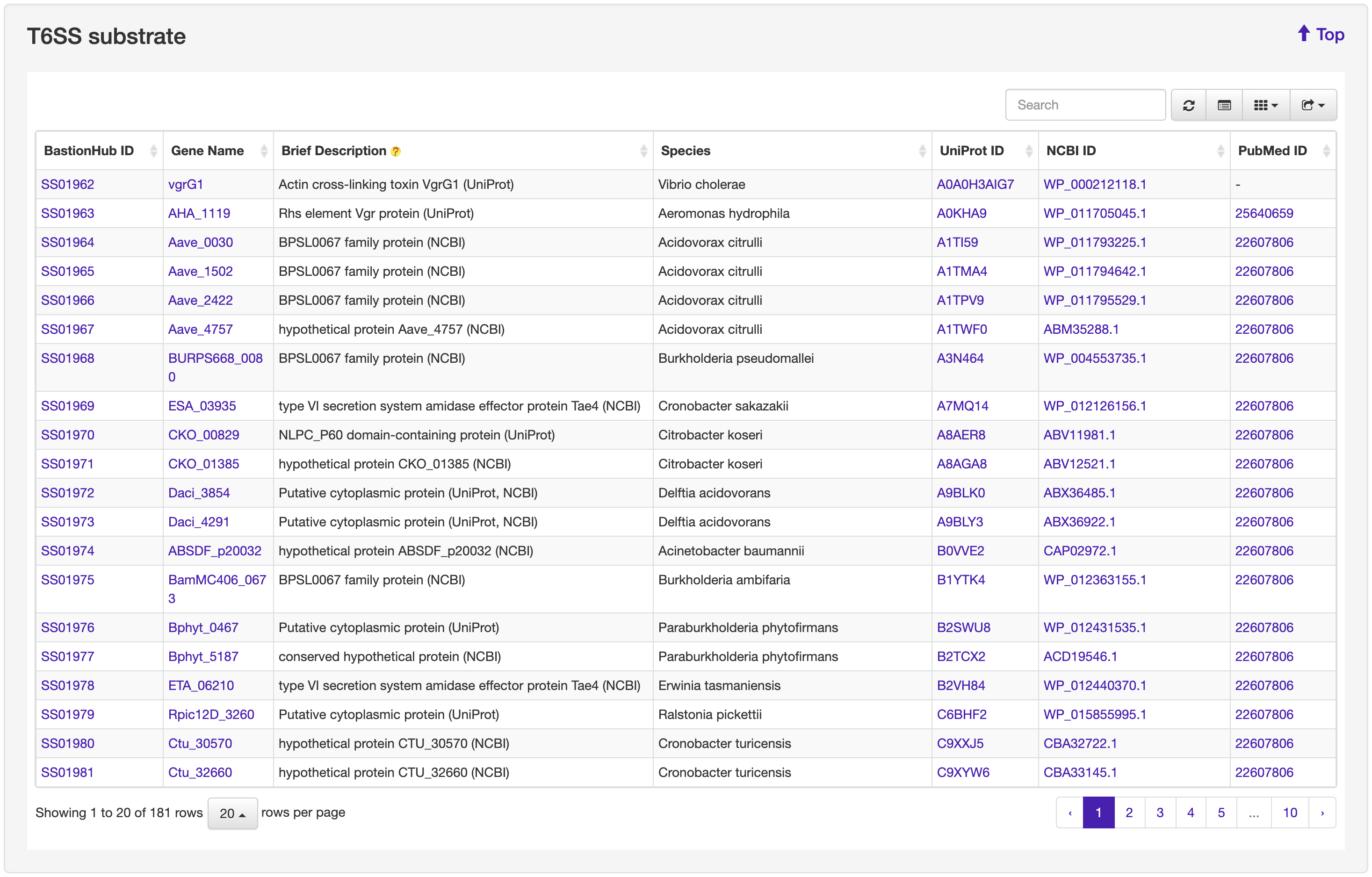
1.3 Search
The Search page of BastionHub provides users with more advanced search options than those available within the Browse page. The search function allows exact queries such as BastionHub, UniProt or NCBI ID, or more broader queries (that don’t require exact matches) using keywords, including protein or gene name and species of origin. We additionally provide a drop-down filter option to further refine results according to features such as conserved domain, protein 3D structure, molecule processing, post-translational modification, metabolic pathway summary, enzymatic and metabolic pathway, mutagenesis, pathogen-host interaction, protein-protein interaction, protein family, or identical protein.

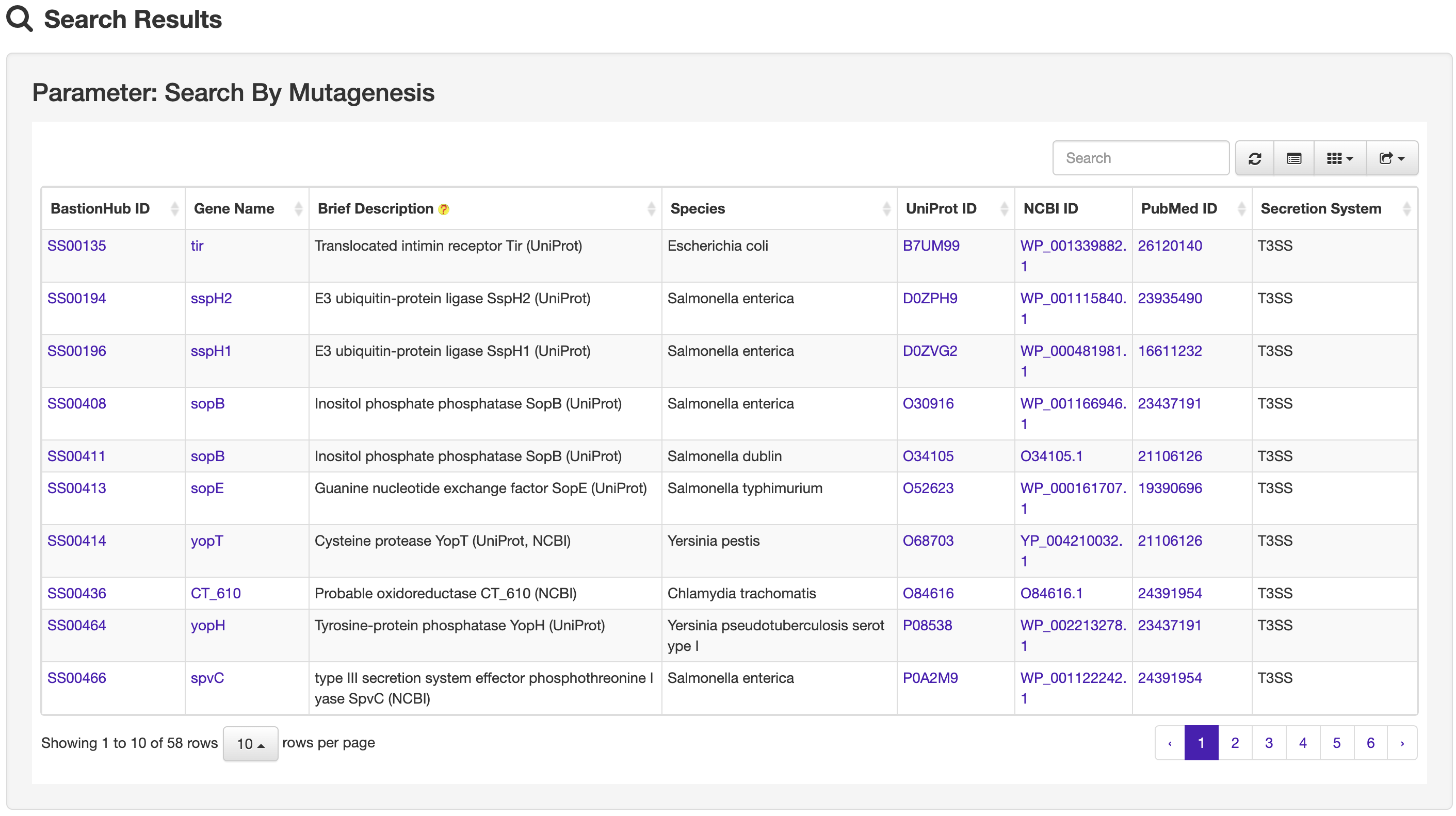
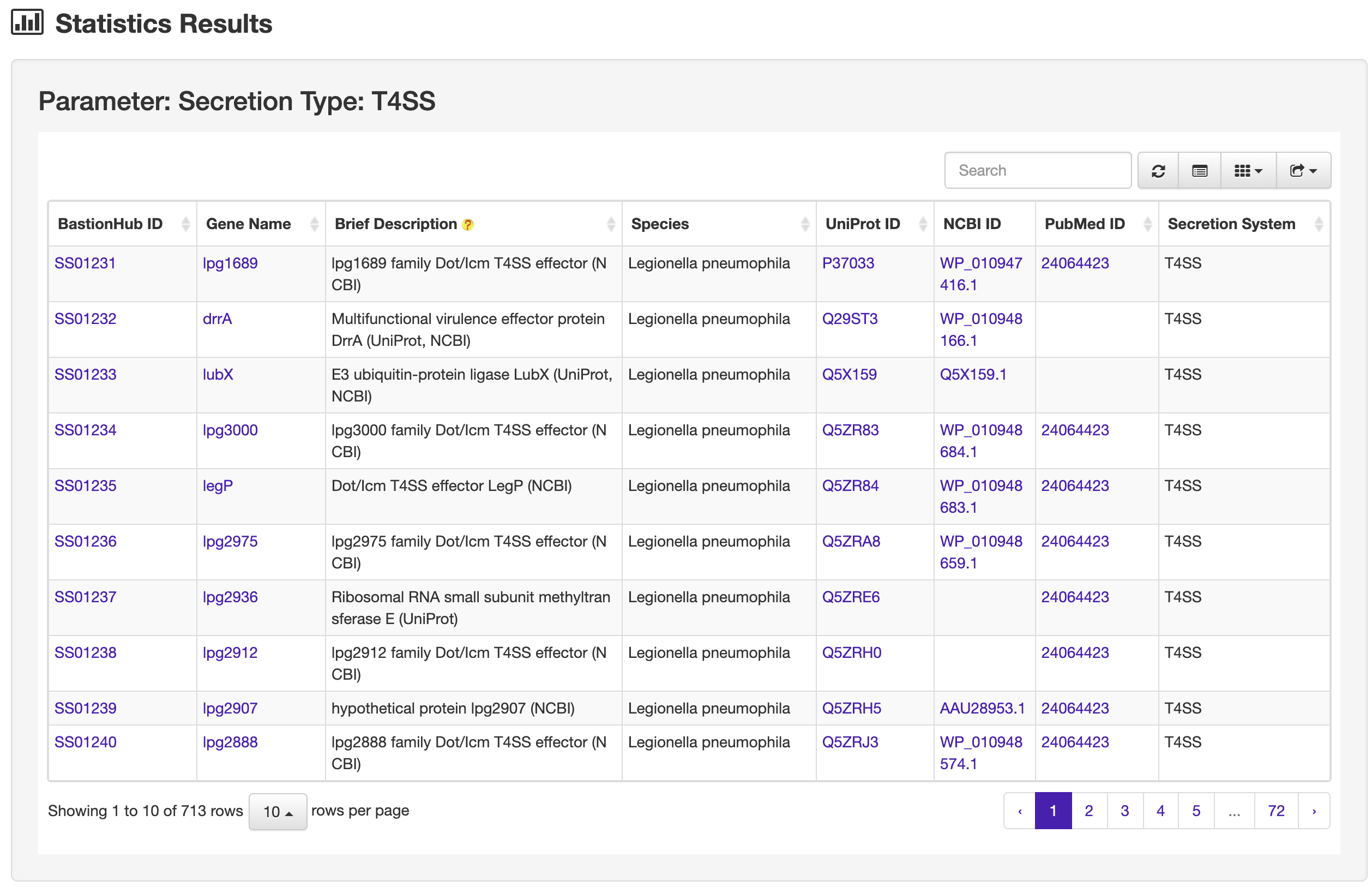
1.4 Statistics
The Statistics page of BastionHub provides multiple options to visualize various type of known substrate proteins, including secretion type distribution, species distribution, phylogenetic tree and homology network.
-
1.4.1 Substrate entries according to secretion type
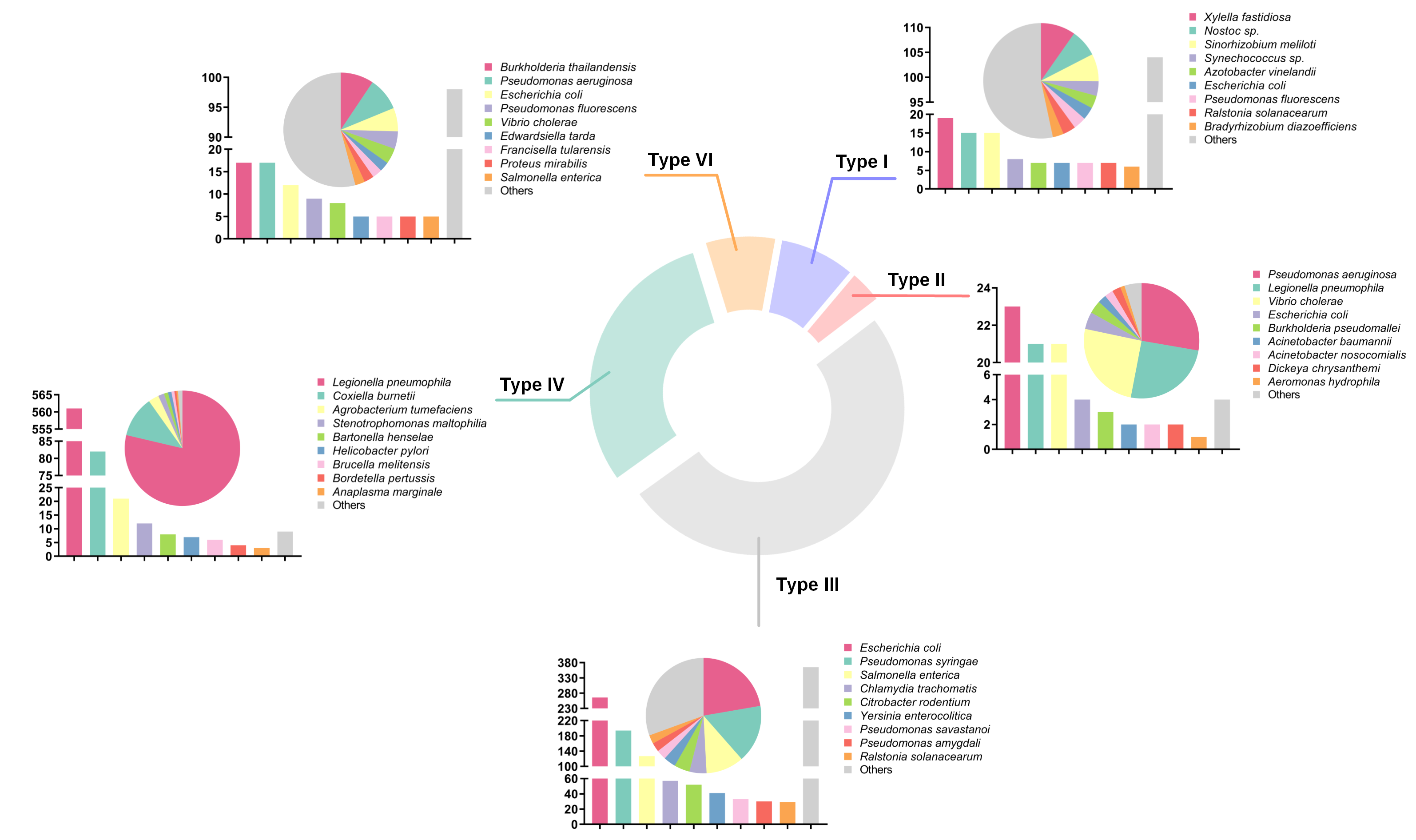
There are five types of secreted substrates in BastionHub: type I secretion system (T1SS) substrates, type II secretion system (T2SS) substrates, type III secretion system (T3SS) substrates, type IV secretion system (T4SS) substrates and type VI secretion system (T6SS) substrates.
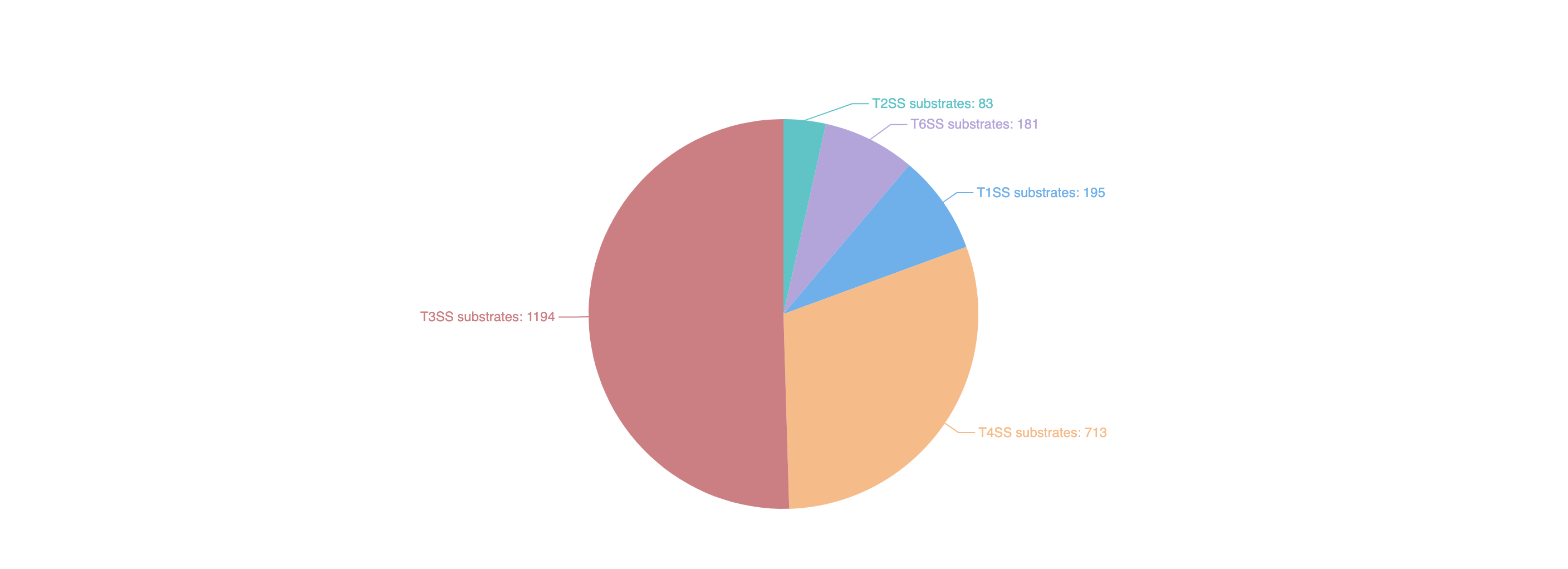
Clicking each section of the pie chart will redirect users to a statistics results page listing the filtered substrates, presented in a similar way to the search results page.
-
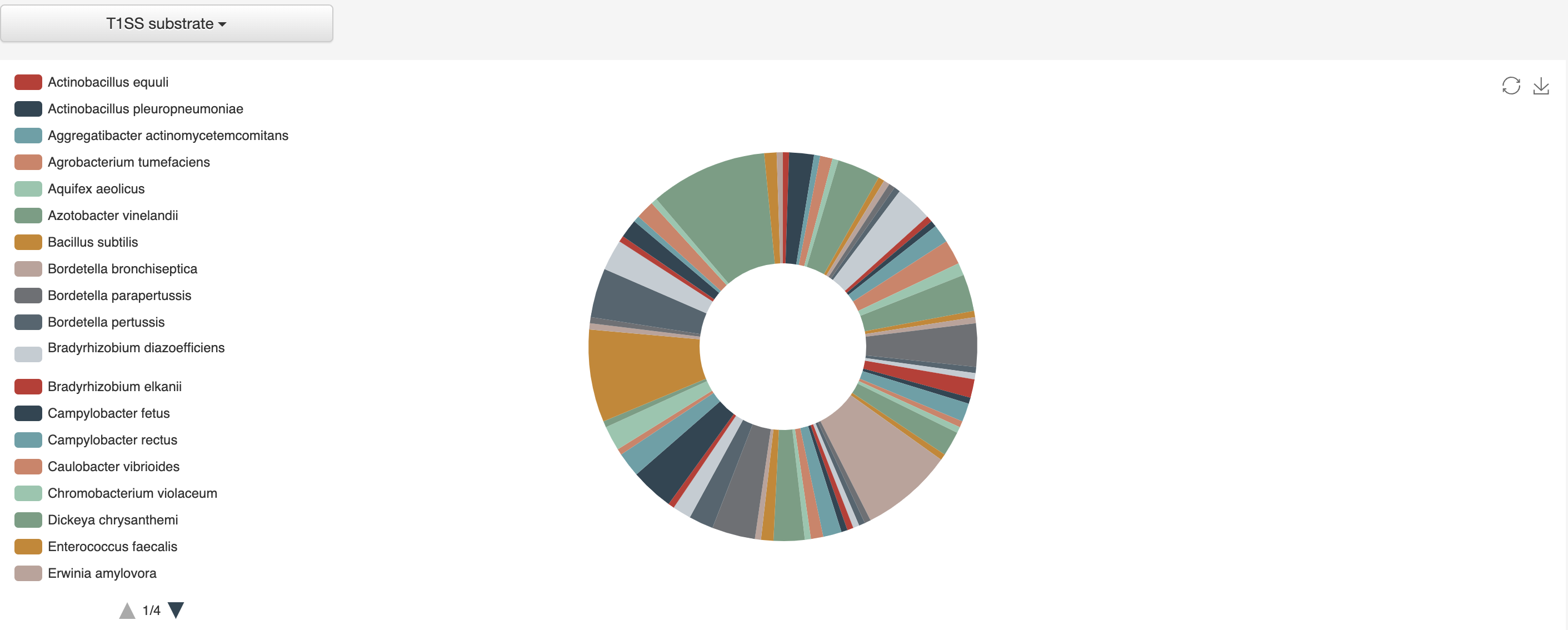
1.4.2 Distribution of substrates according to bacterial species
-
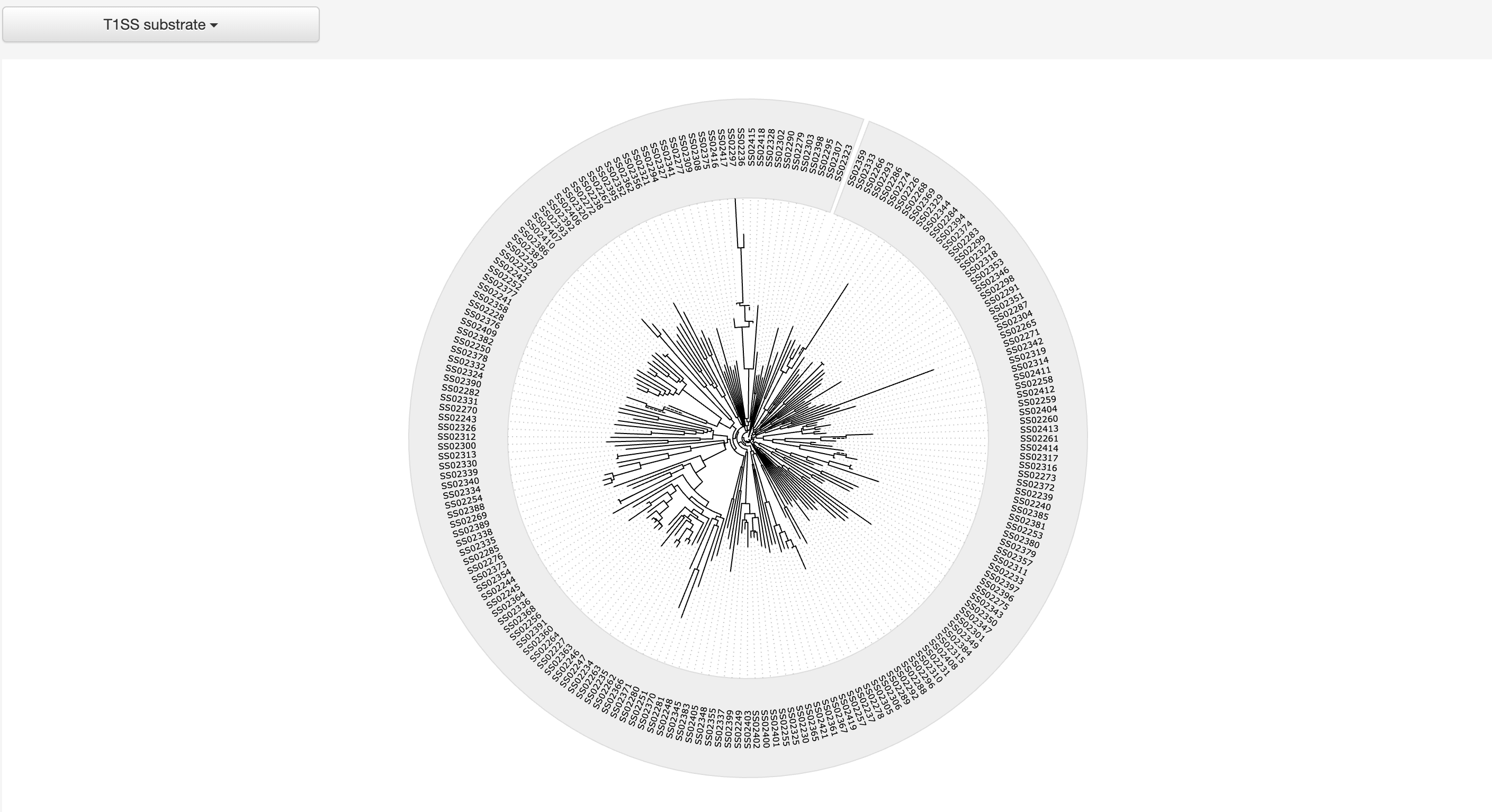
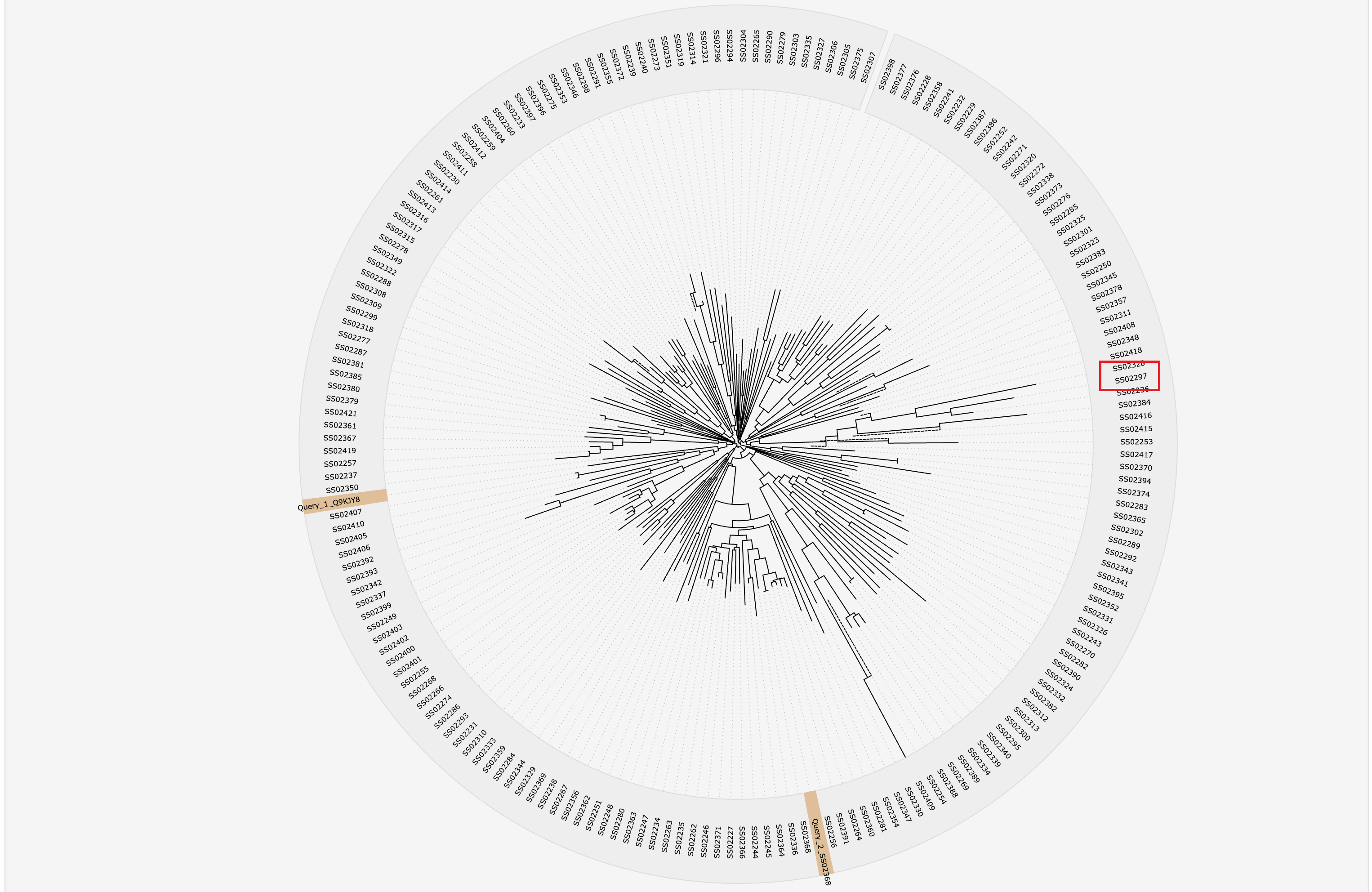
1.4.3 Phylogenetic tree of various types of substrates
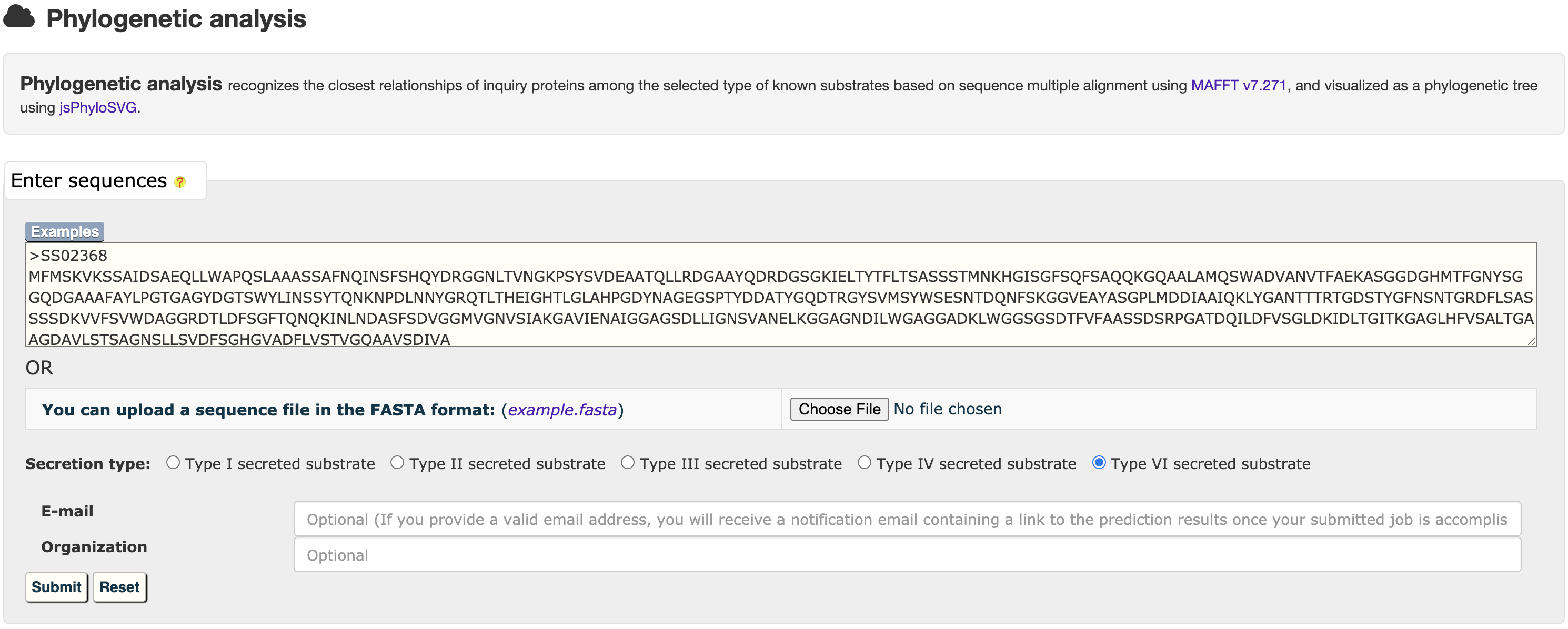
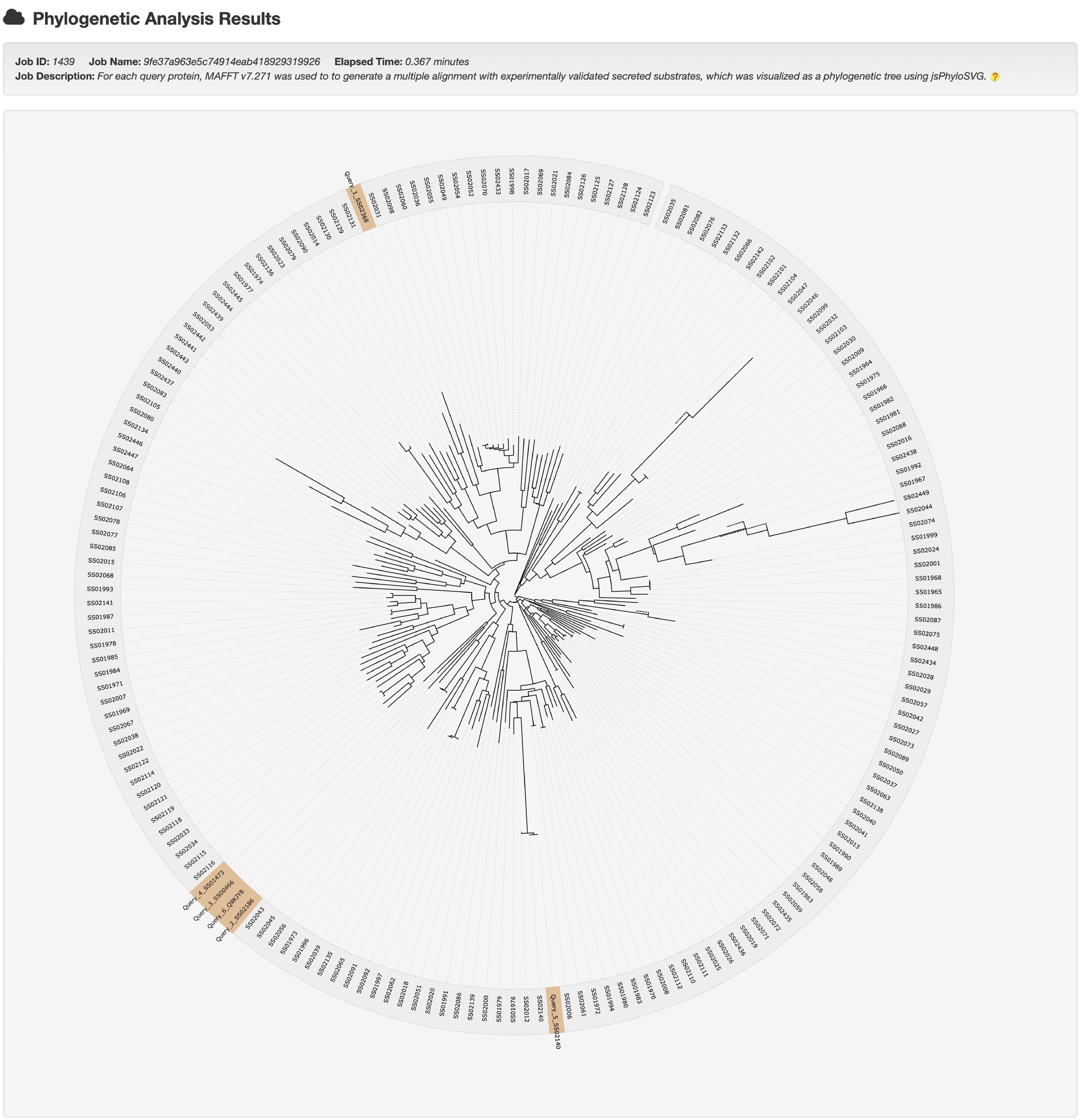
The MAFFT v7.271 was used to generate multiple alignment results among selected type of known substrate proteins, which was visualized by jsPhyloSVG.
-
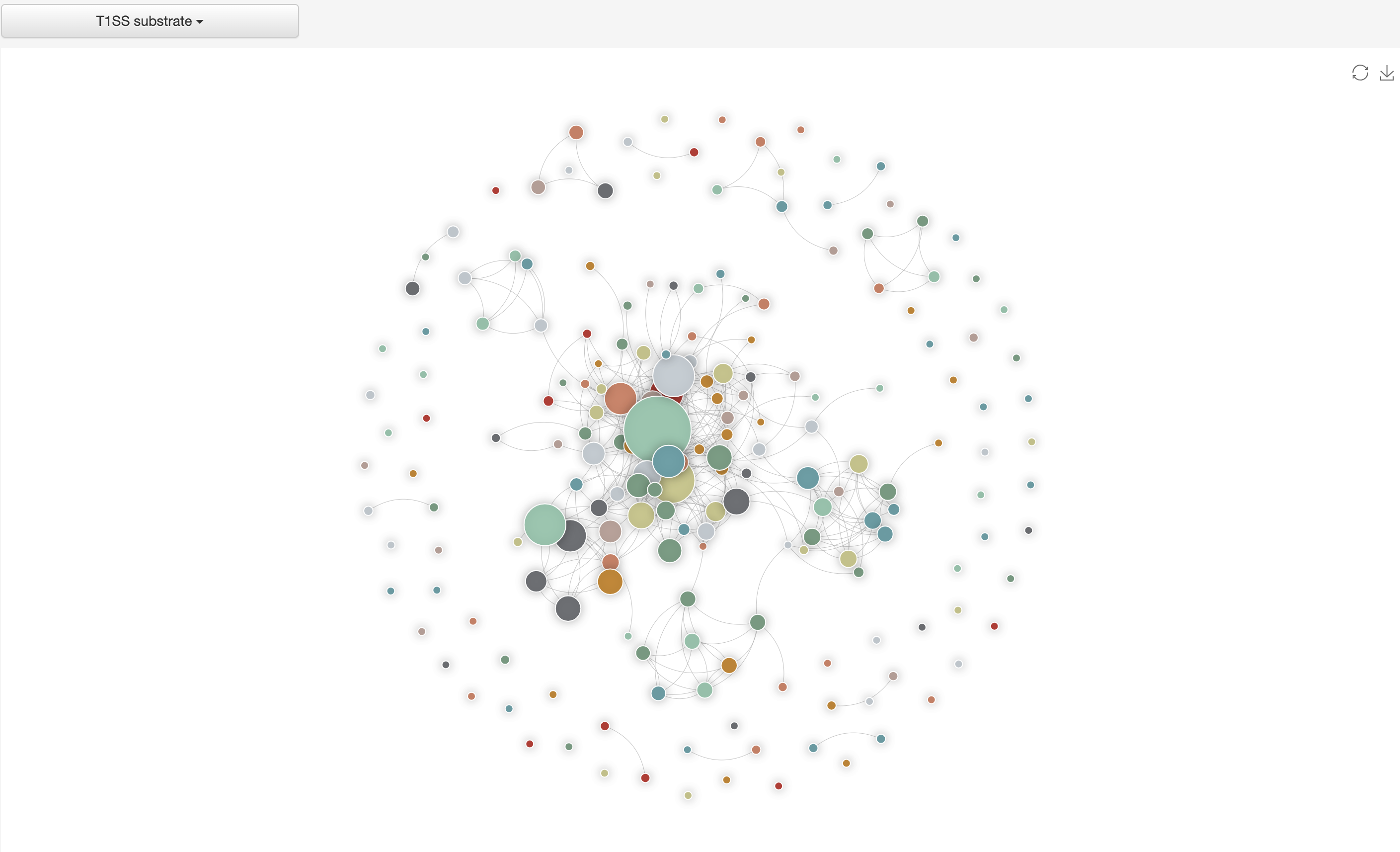
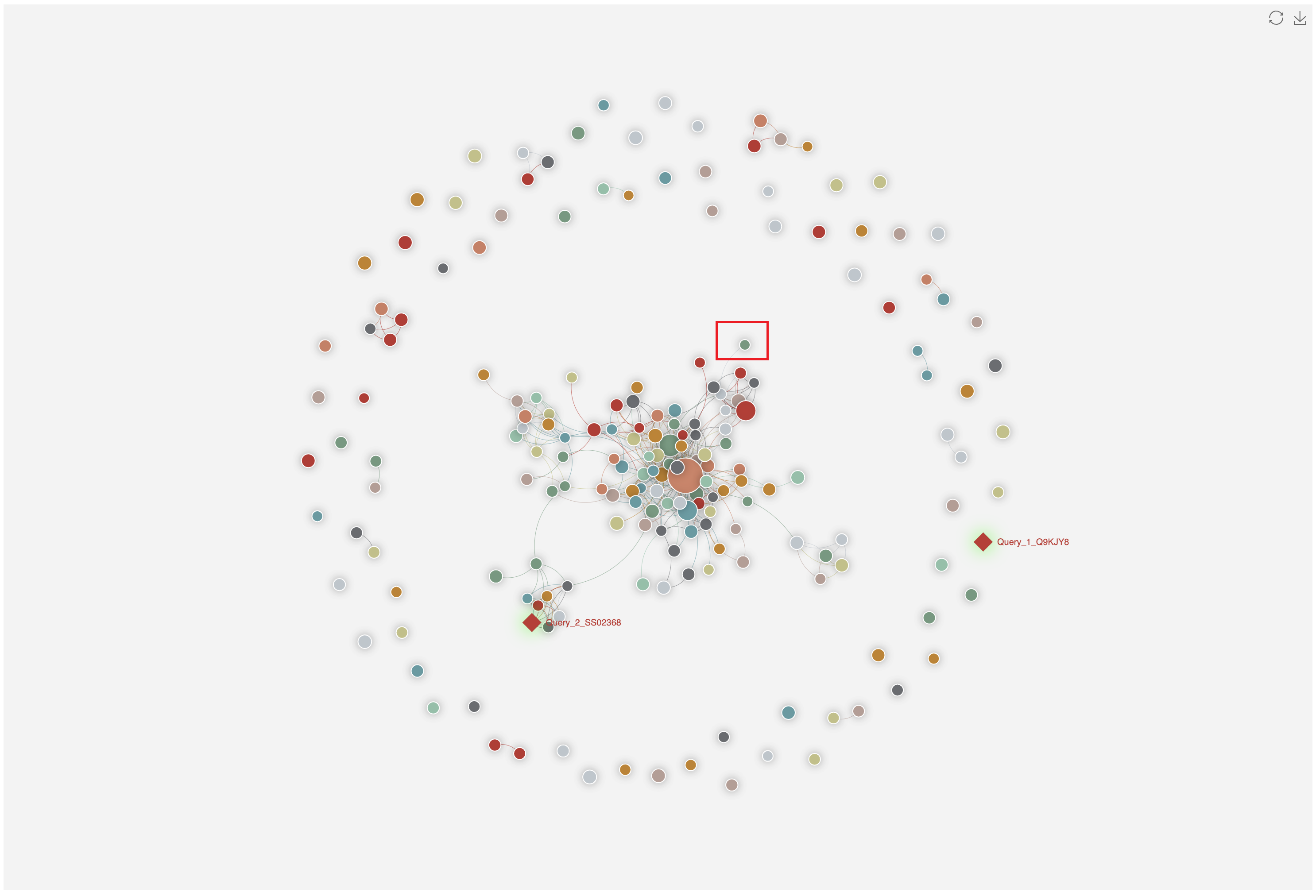
1.4.4 Homology network of various types of substrates
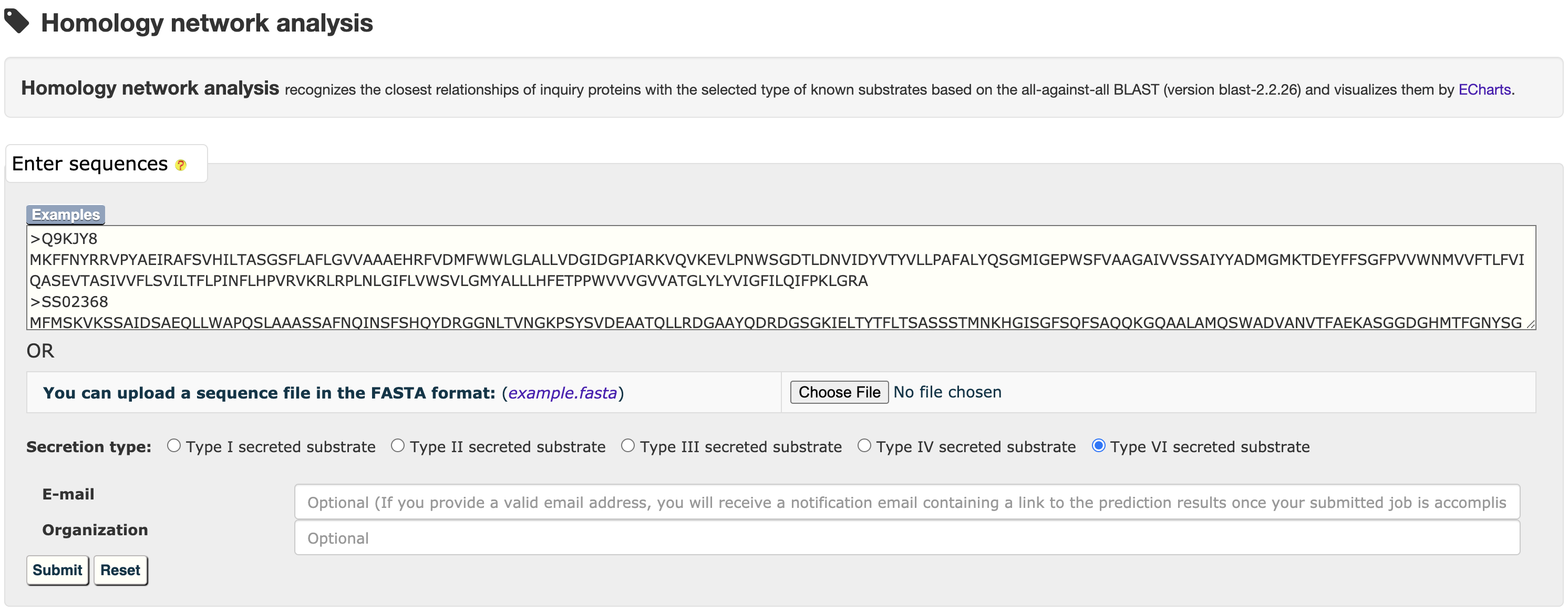
The all-against-all BLAST (version blast-2.2.26) was used to generate the sequence similarity network among the selected type of substrates, which was visualized by ECharts.
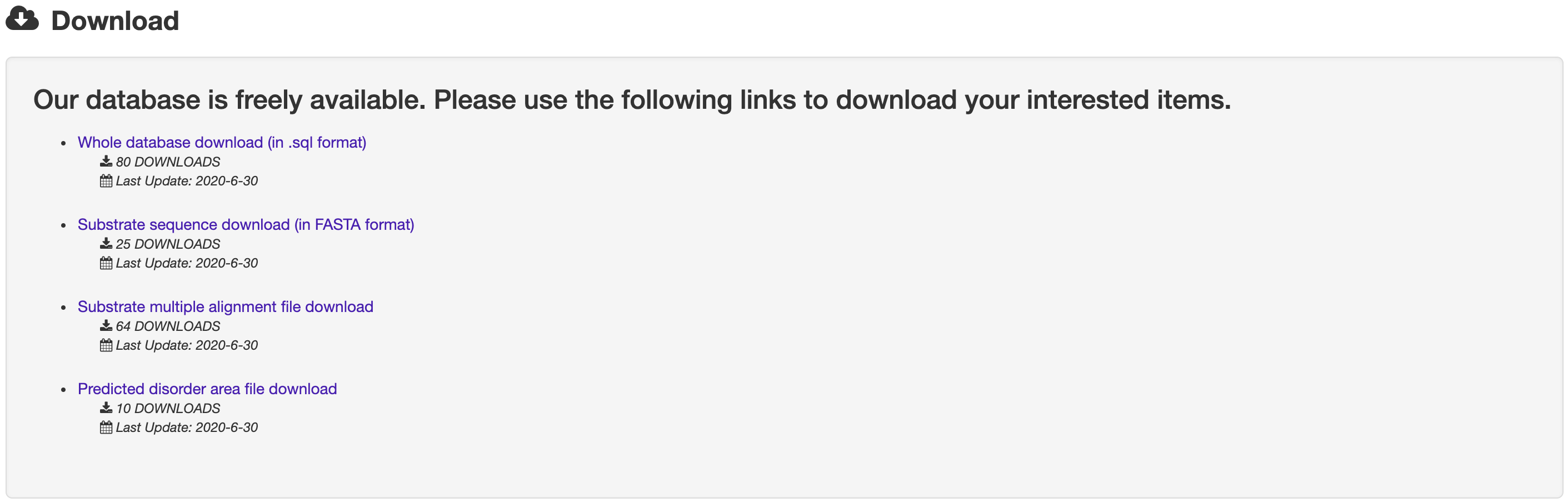
1.5 Download
The Download page of BastionHub provides multiple options for users to to download files, including the whole database (in sql format), sequences (in FASTA format), disorder files and multiple sequence alignments.
1.6 Detailed information
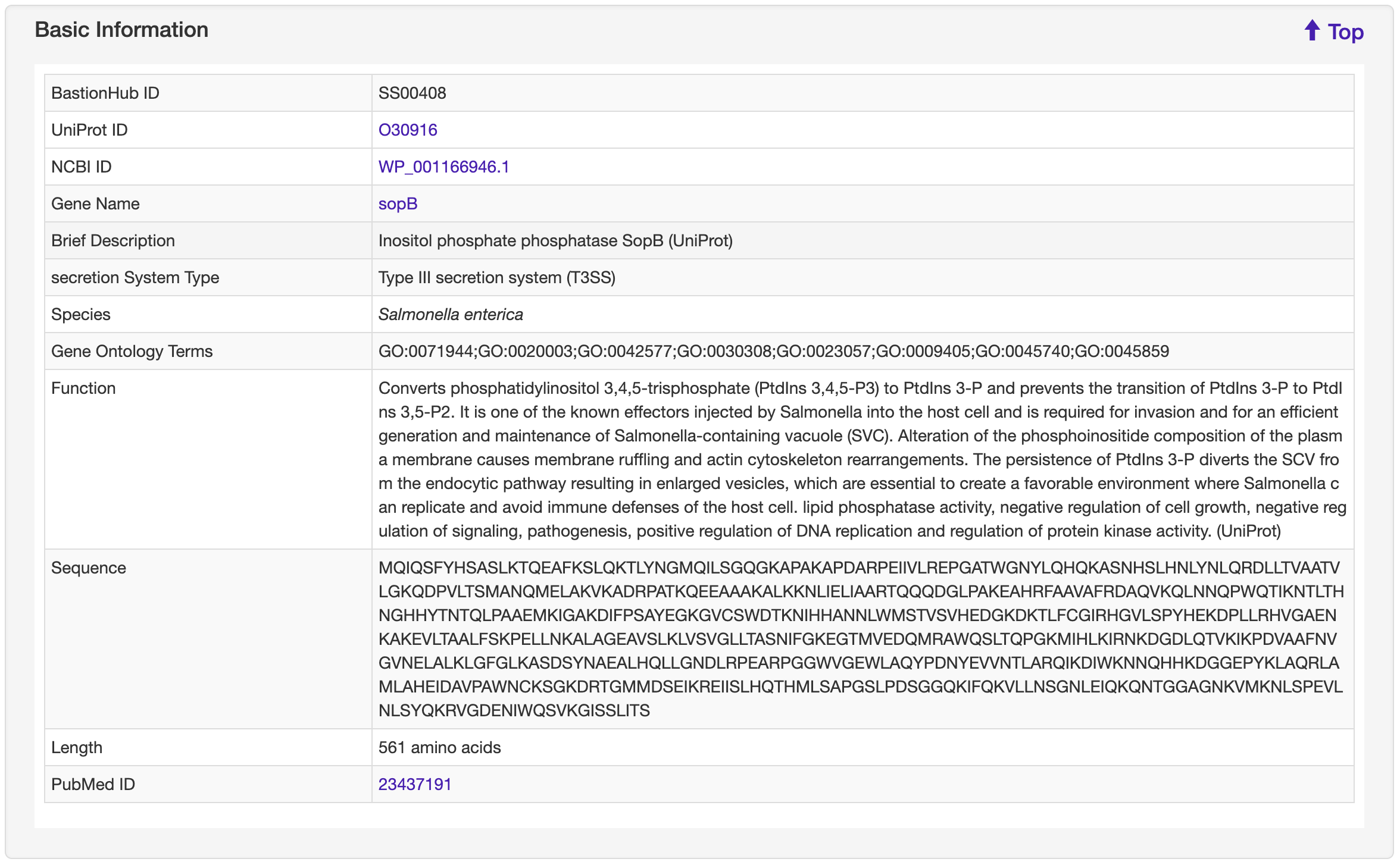
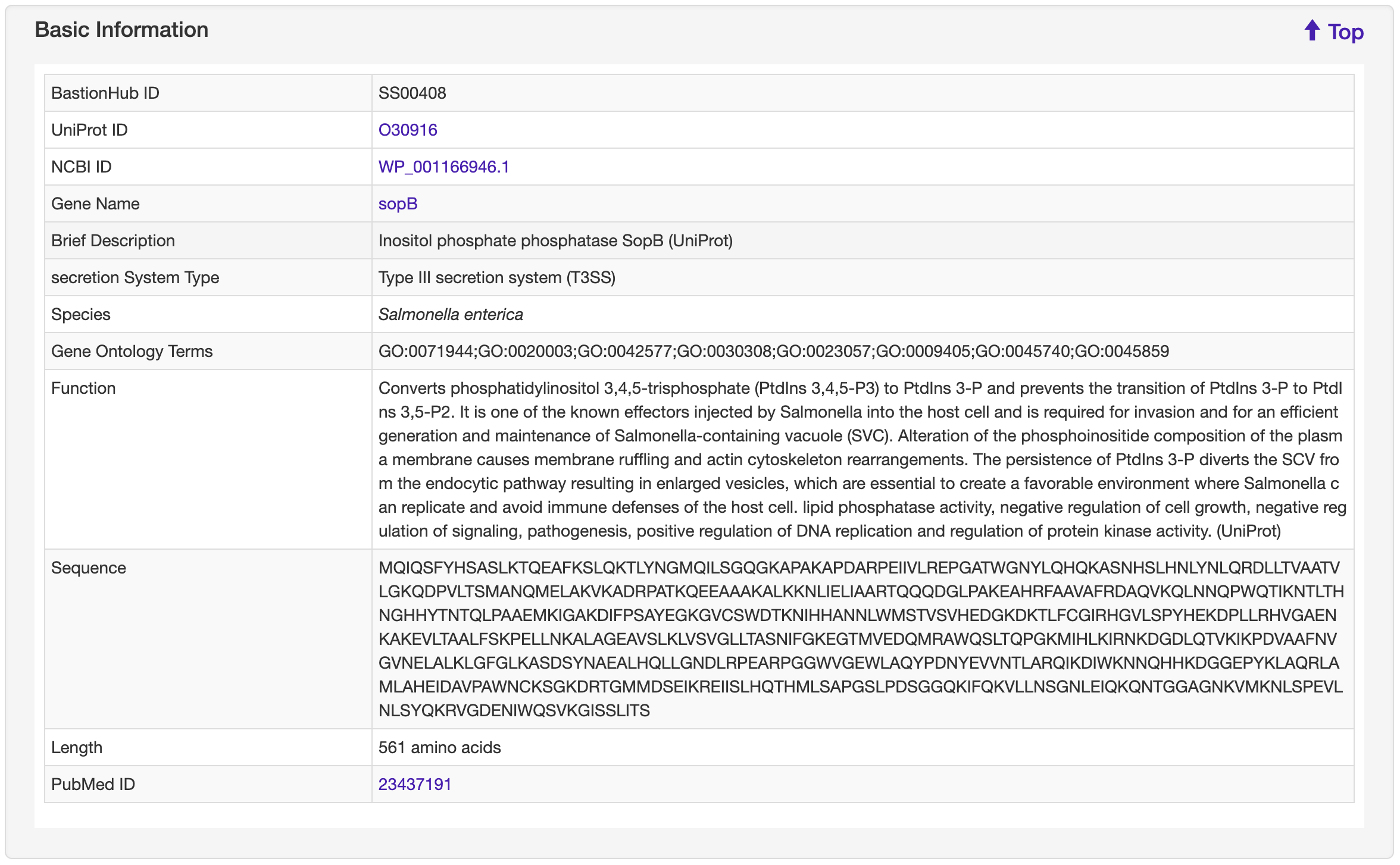
The Detailed information page provides detailed annotations for each substrate comprising their basic information, advanced annotations, and relationship analyses among their associated type of known substrates. Basic information consists of their UniProt ID, NCBI ID, gene name, brief description, secretion system type, species, gene ontology terms, function, sequence, length, and PubMed ID. For advanced annotations, we incorporated conserved domains depicted on 2D protein maps, interactive 3D protein structures, predicted disorder area, molecule processing and post-translational modification information, metabolic pathway summaries, enzymatic and metabolic pathway details, mutagenesis results, pathogen-host Interactions, protein-protein interactions and protein families. Finally, we included five pre-calculated relationship analyses for each substrate: lists of 100% identical proteins indexed by BastionHub that would normally be consolidated into a single entry, but based on their different species, annotations or sources, were kept as individual entries, and similar proteins within BastionHub (if available), multiple sequence alignments, a phylogenetic tree, and a homology network.
-
1.6.1 Basic Information
-
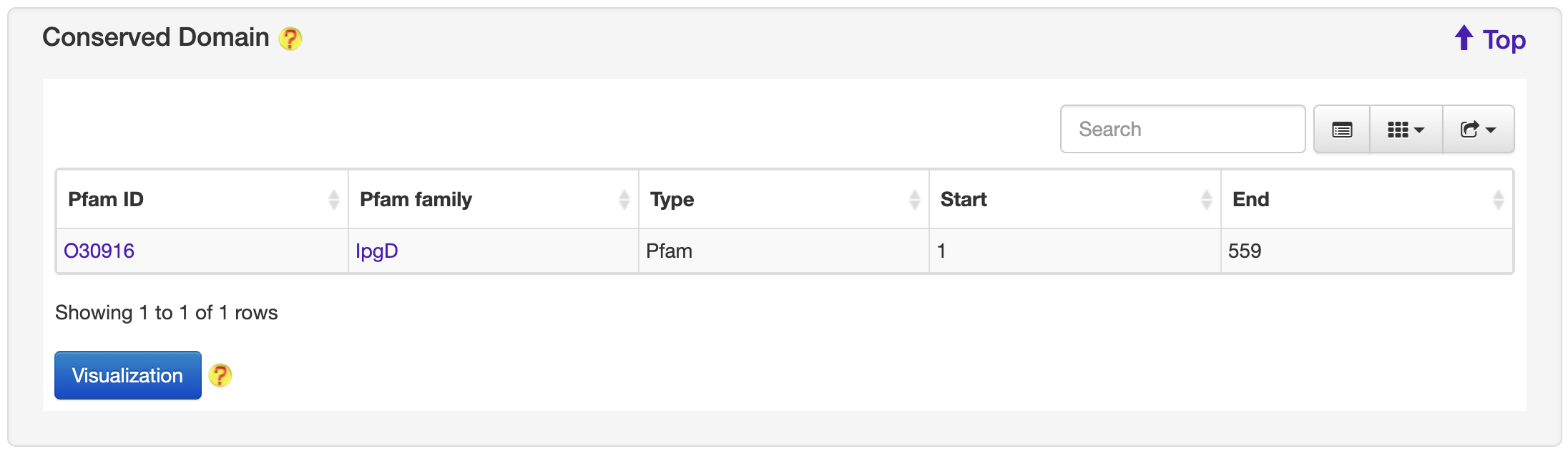
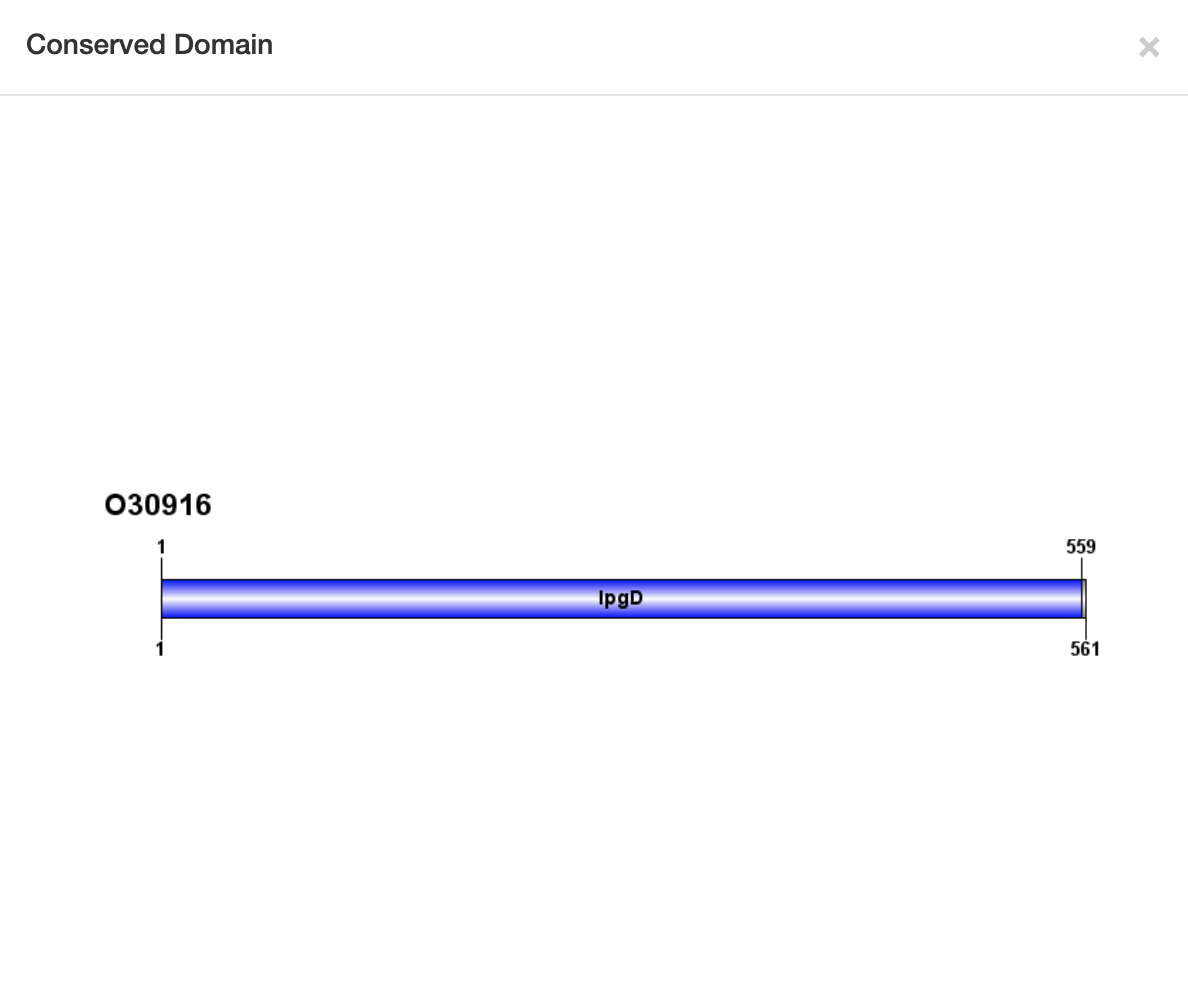
1.6.2 Conserved Domain
For each entry, the Conserved Domain where available was collected from the Pfam database.
-
1.6.3 Protein 3D Structure
For each entry, the Protein 3D Structure information where available was collected from the PDB database. A 3-D visualization was provided by using PDB LiteMol. Following is an example for SS00408.
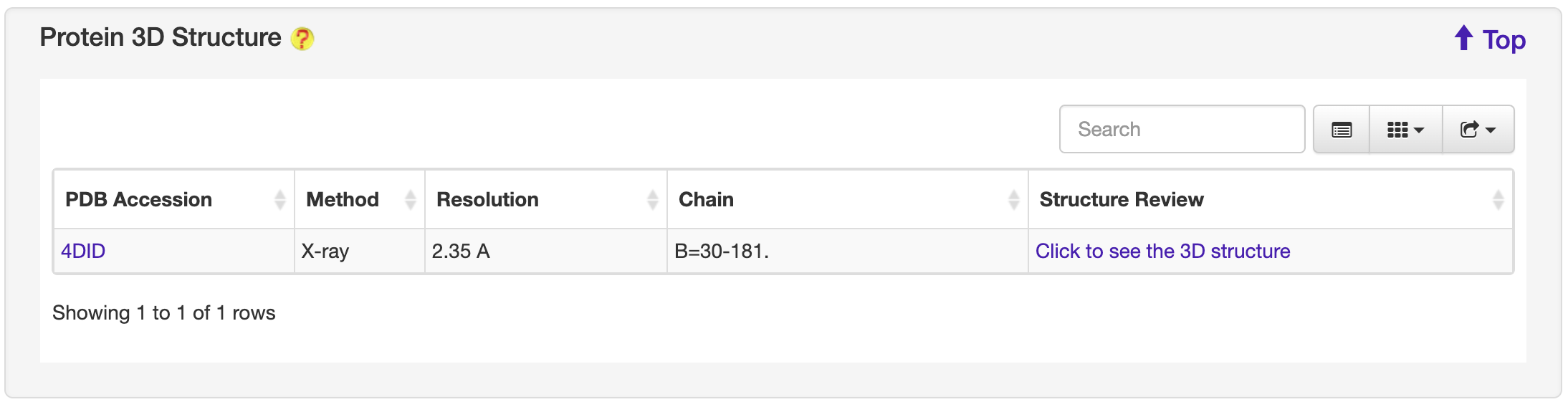
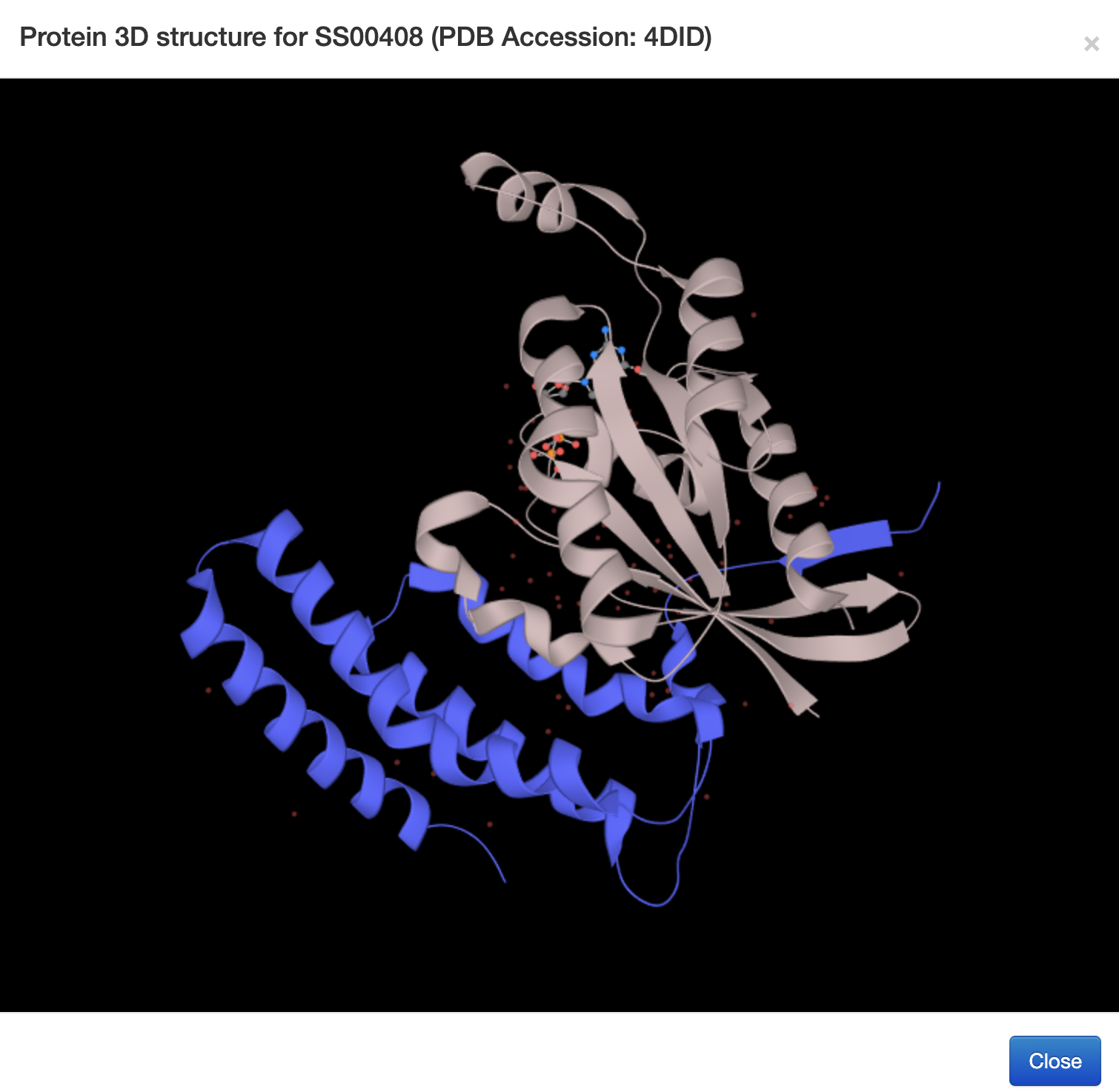
-
1.6.4 Disorder Area
For each entry, the Disorder Area was generated by the IUPred2A server, and visualized by ECharts.
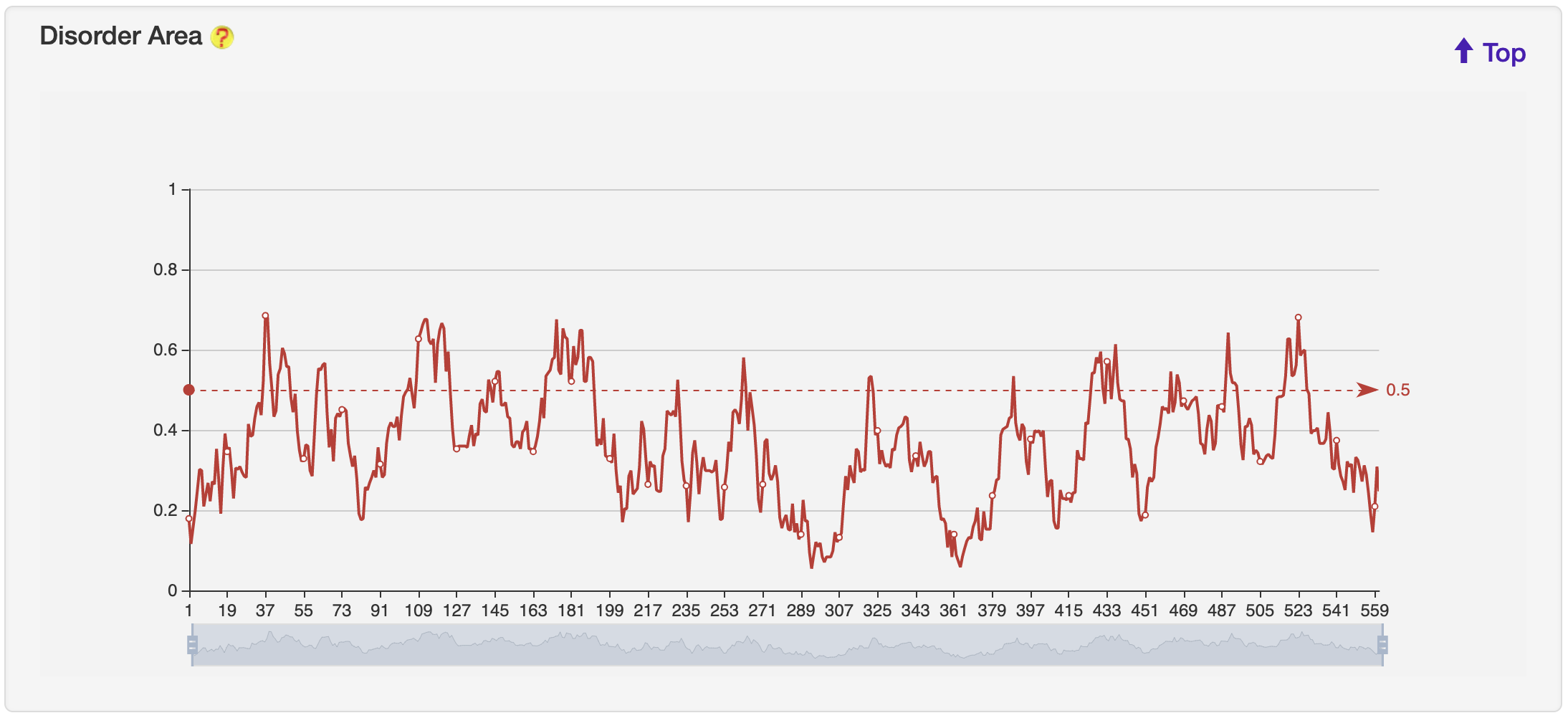
-
1.6.5 Molecule Processing
For each entry, the Molecule Processing information where available was collected from the UniProt database.
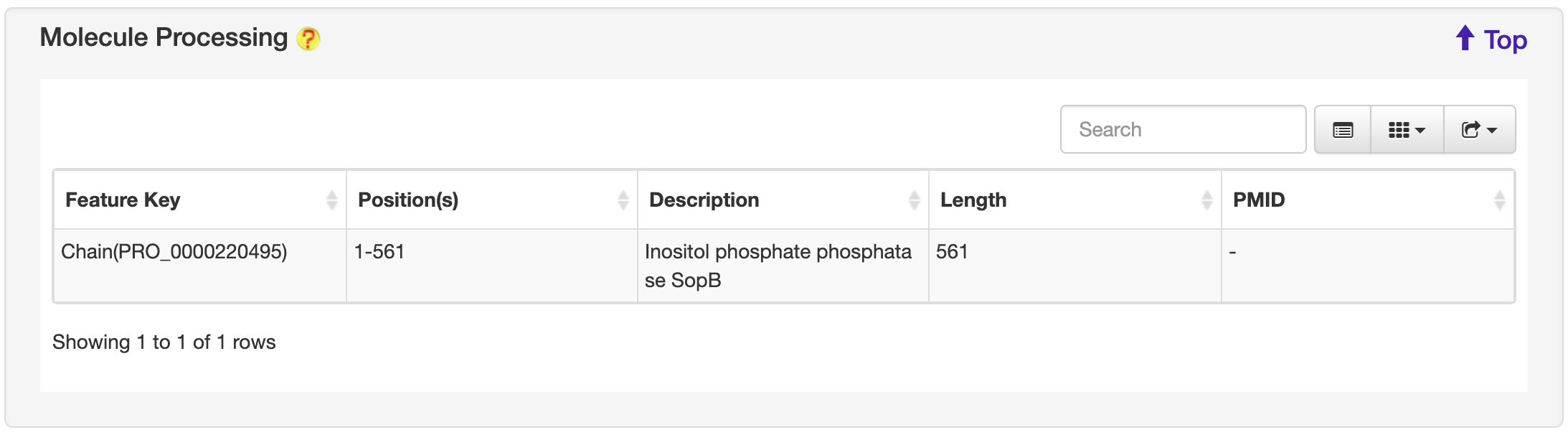
-
1.6.6 Post-translational Modification
For each entry, the Post-translational Modification information where available was collected from the UniProt database.

-
1.6.7 Metabolic Pathway Summary
For each entry, the Metabolic Pathway Summary information where available was collected from the UniProt database.

-
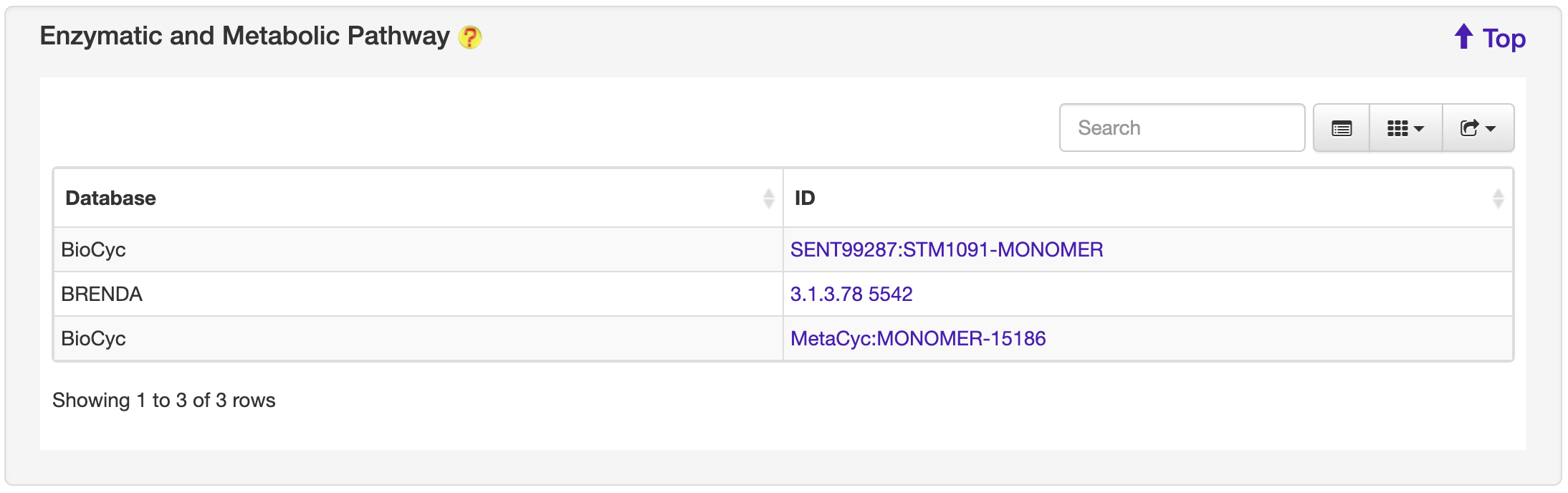
1.6.8 Enzymatic and Metabolic Pathway
For each entry, the Enzymatic and Metabolic Pathway information where available was collected from the BioCyc and BRENDA database.
-
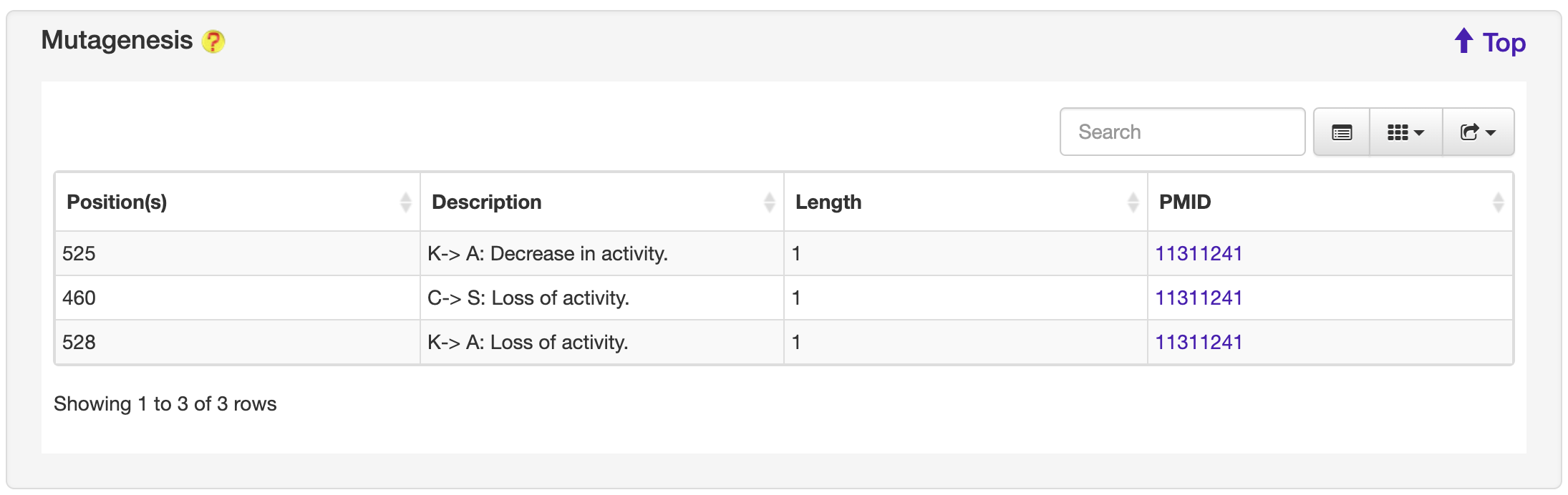
1.6.9 Mutagenesis
For each entry, the Mutagenesis information where available was collected from the UniProt database.
-
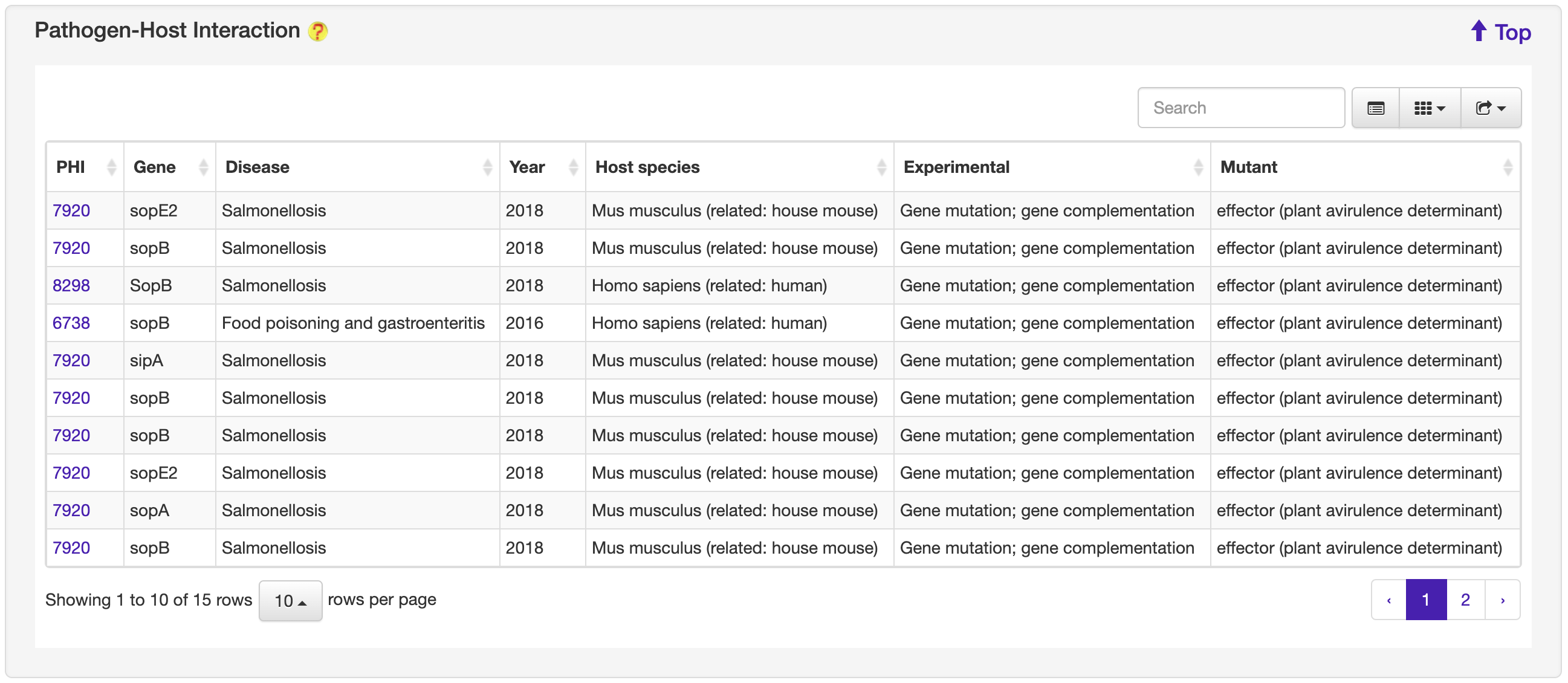
1.6.10 Pathogen-Host Interaction
For each entry, the Pathogen-Host Interaction information where available was collected from the PHI-base database.
-
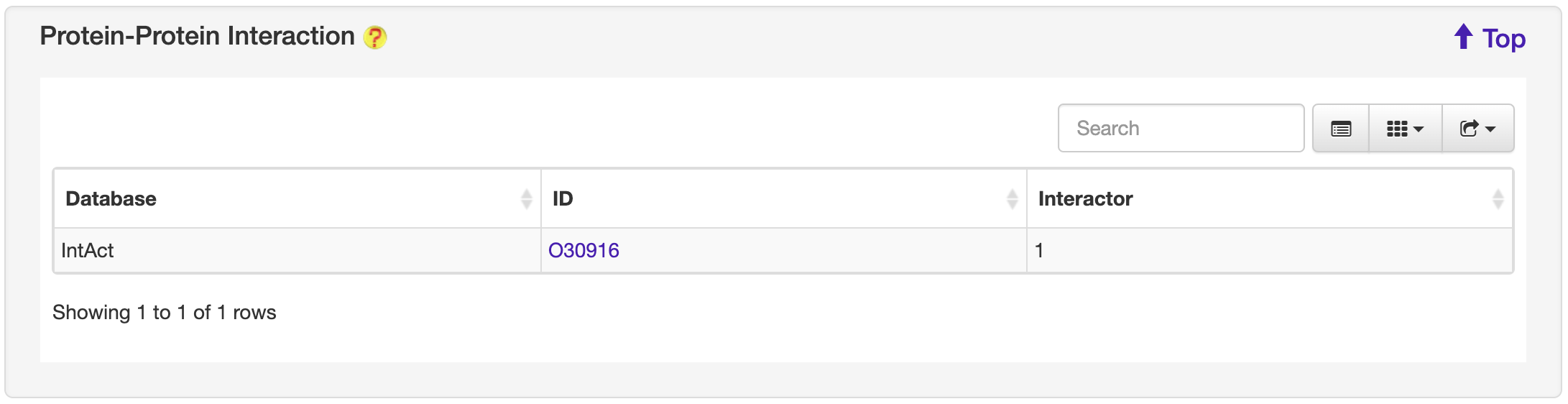
1.6.11 Protein-Protein Interaction
For each entry, the Protein-Protein Interaction information where available was collected from the STRING, DIP, IntAct and MINT database.
-
1.6.12 Protein Family
For each entry, the Protein Family information where available was collected from the UniProt database.

-
1.6.13 Identical Protein
For each entry, the Identical Protein information where available in the BastionHub database was presented.
-
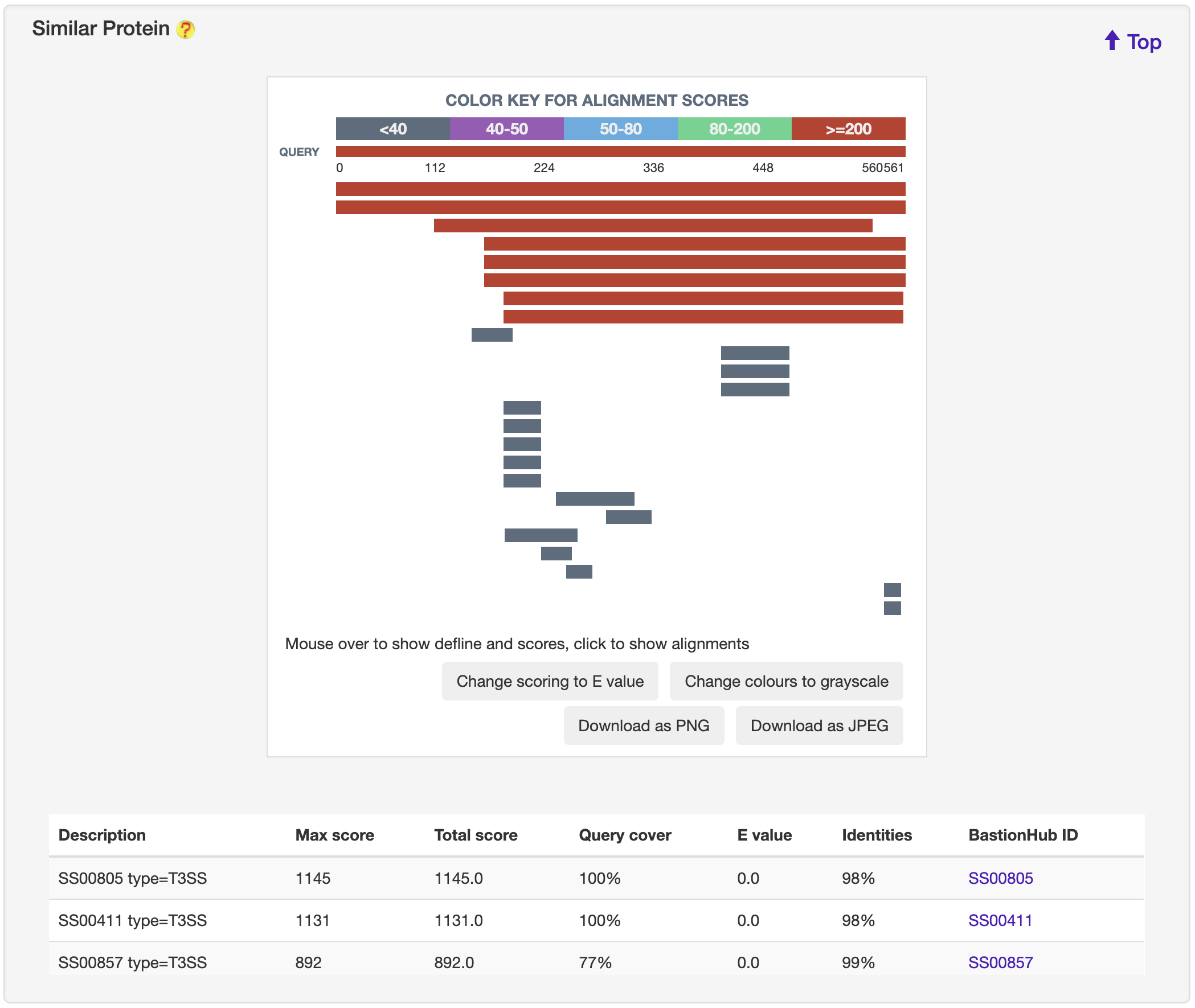
1.6.14 Similar Protein
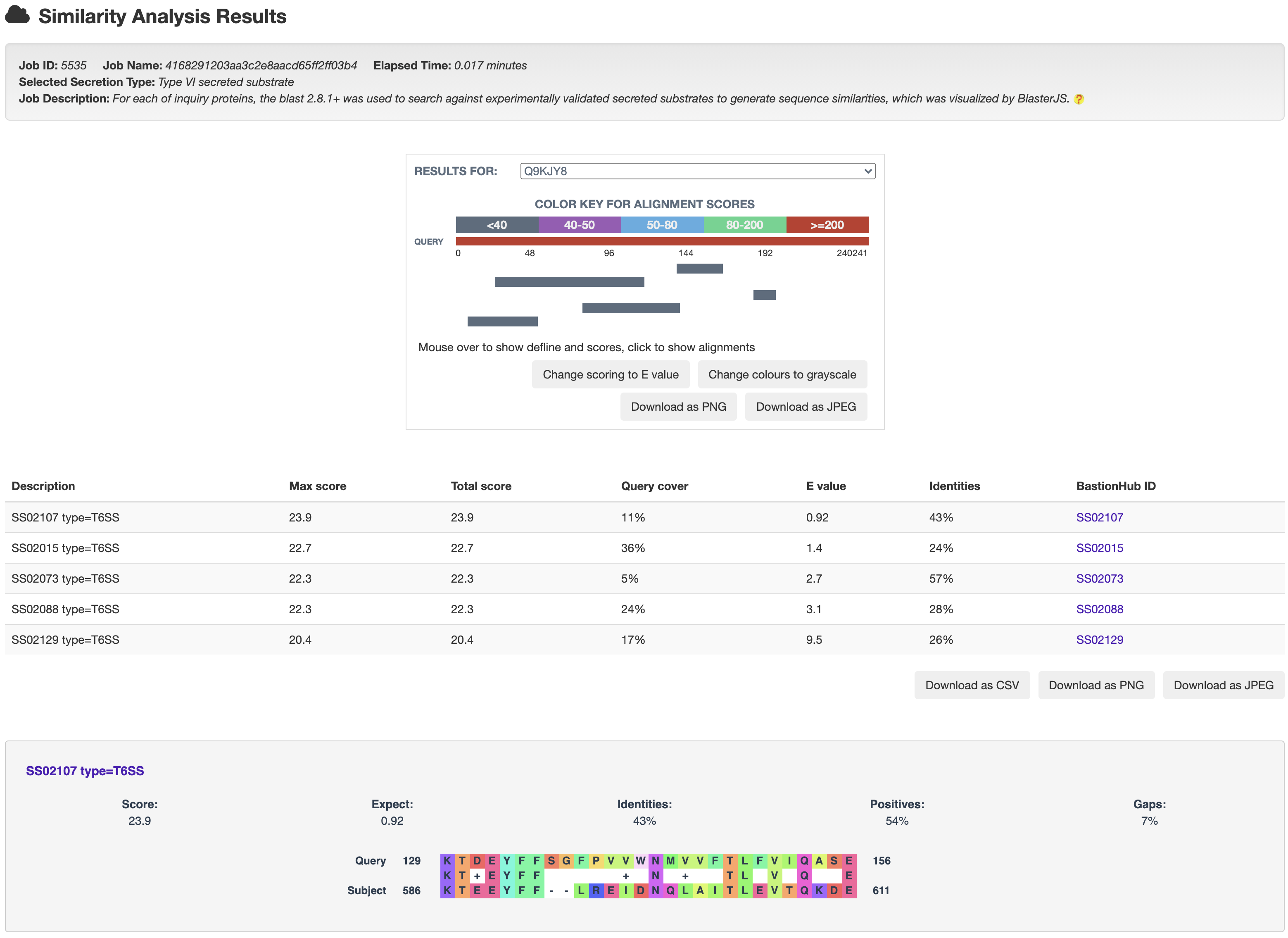
For each entry, the blast 2.8.1+ was used to search against its associated type of known substrates to generate sequence similarities, which was visualized by BlasterJS.
-
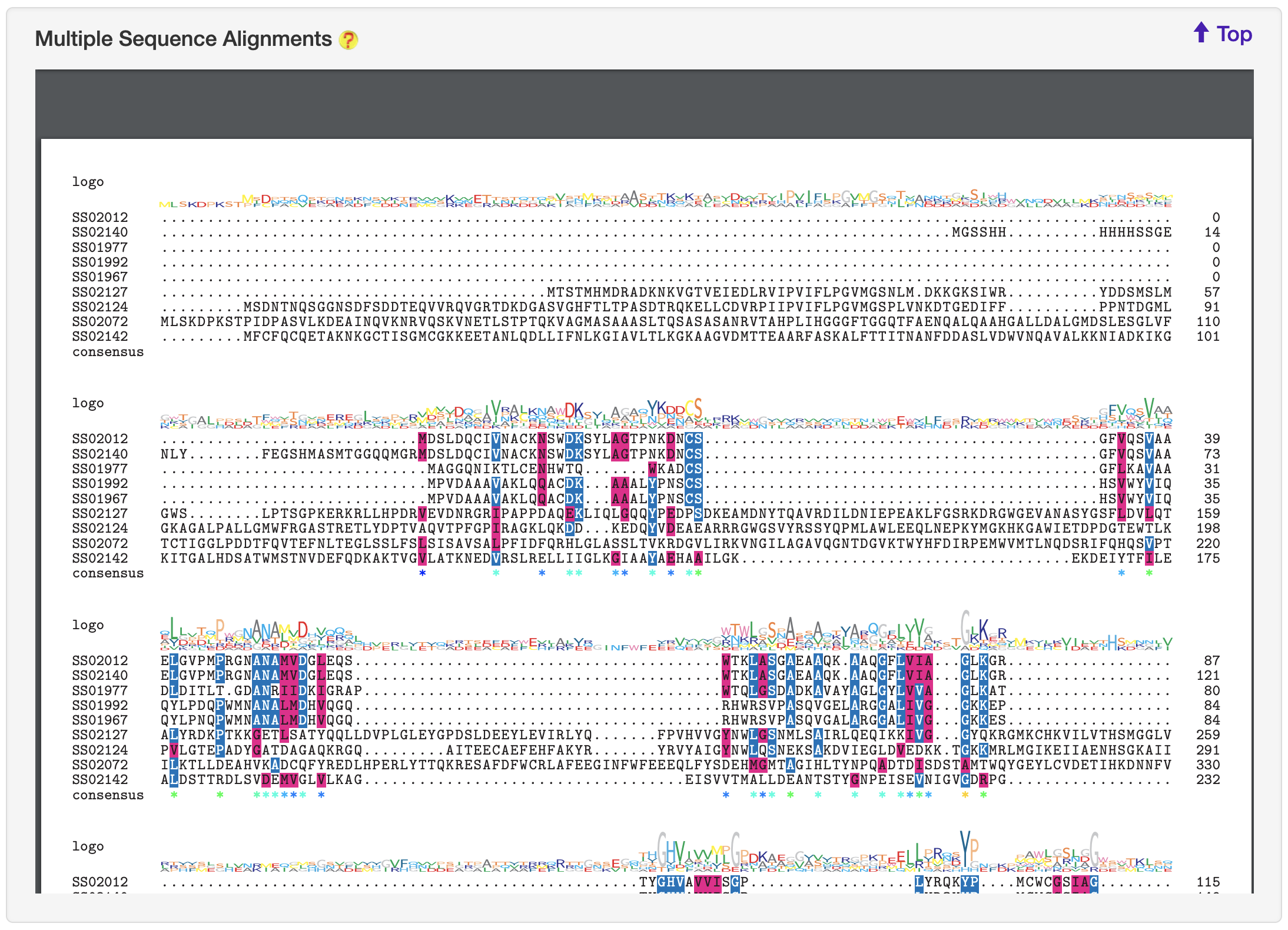
1.6.15 Multiple Sequence Alignments
For each entry, blast-2.8.1+ was used to search against its associated type of known substrates to obtain the homologous sequences. This entry with its retrieved homologous sequences was used to generate multiple alignment file (executed by ClustalW but invoked by msa) and then visualized by the the R library msa.
-
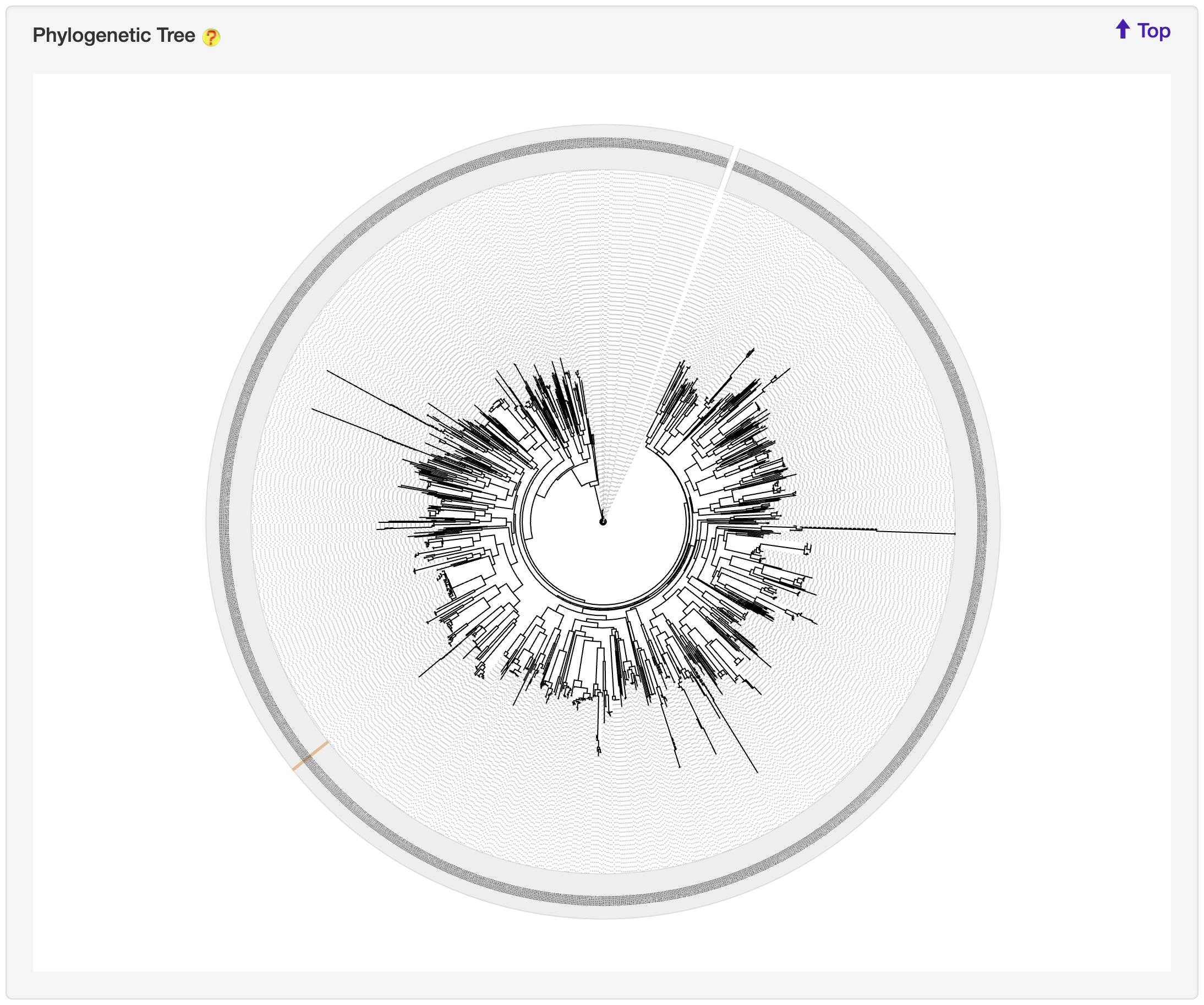
1.6.16 Phylogenetic Tree
For each entry, the MAFFT v7.271 was used to generate multiple alignment results against its associated type of known substrates, which was visualized by jsPhyloSVG.
-
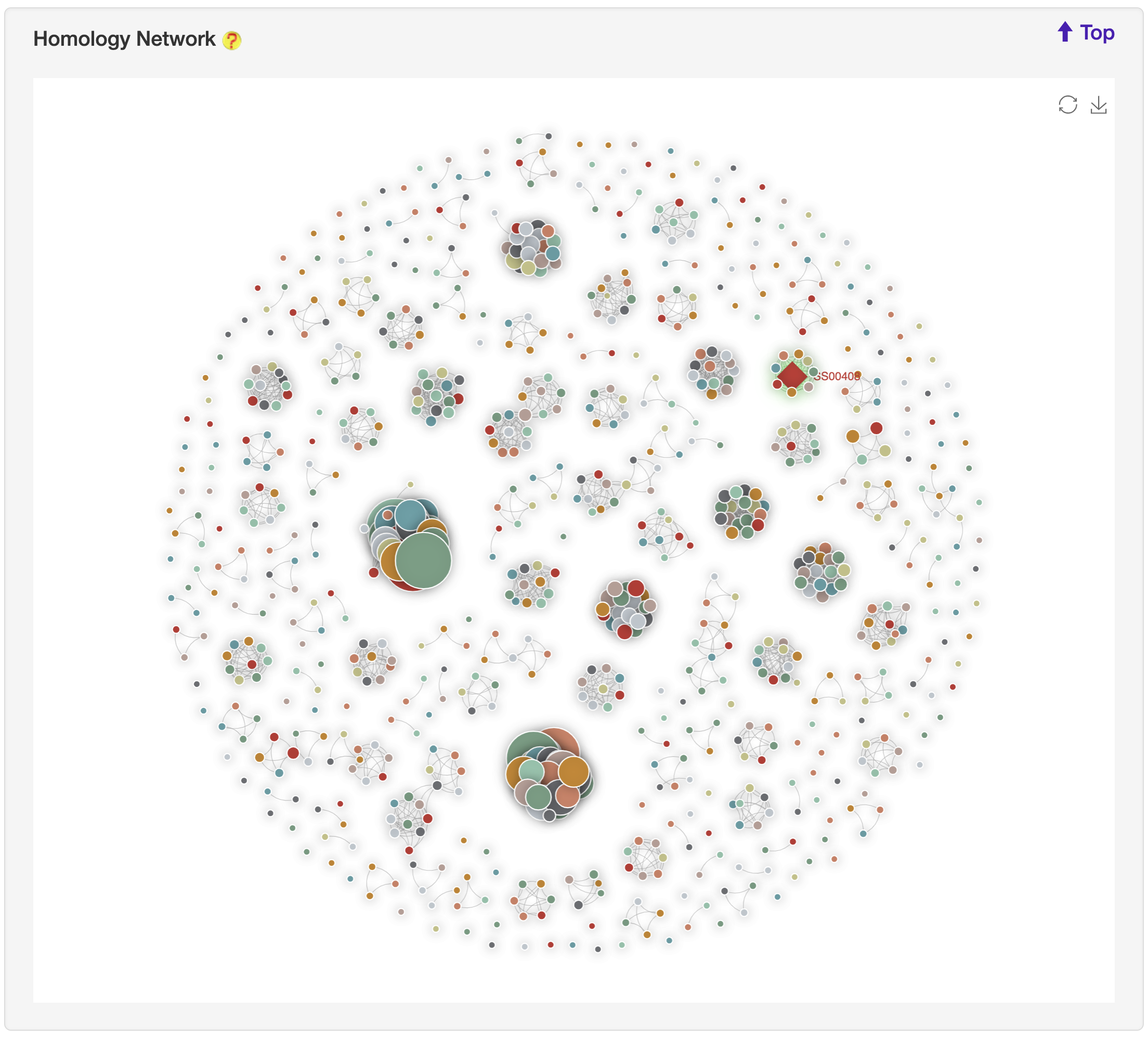
1.6.17 Homology Network
For each entry (represented by red rhombus), the all-against-all BLAST (version blast-2.2.26) was used upon itself and its associated type of known substrates to generate the sequence similarity network, visualized by ECharts.
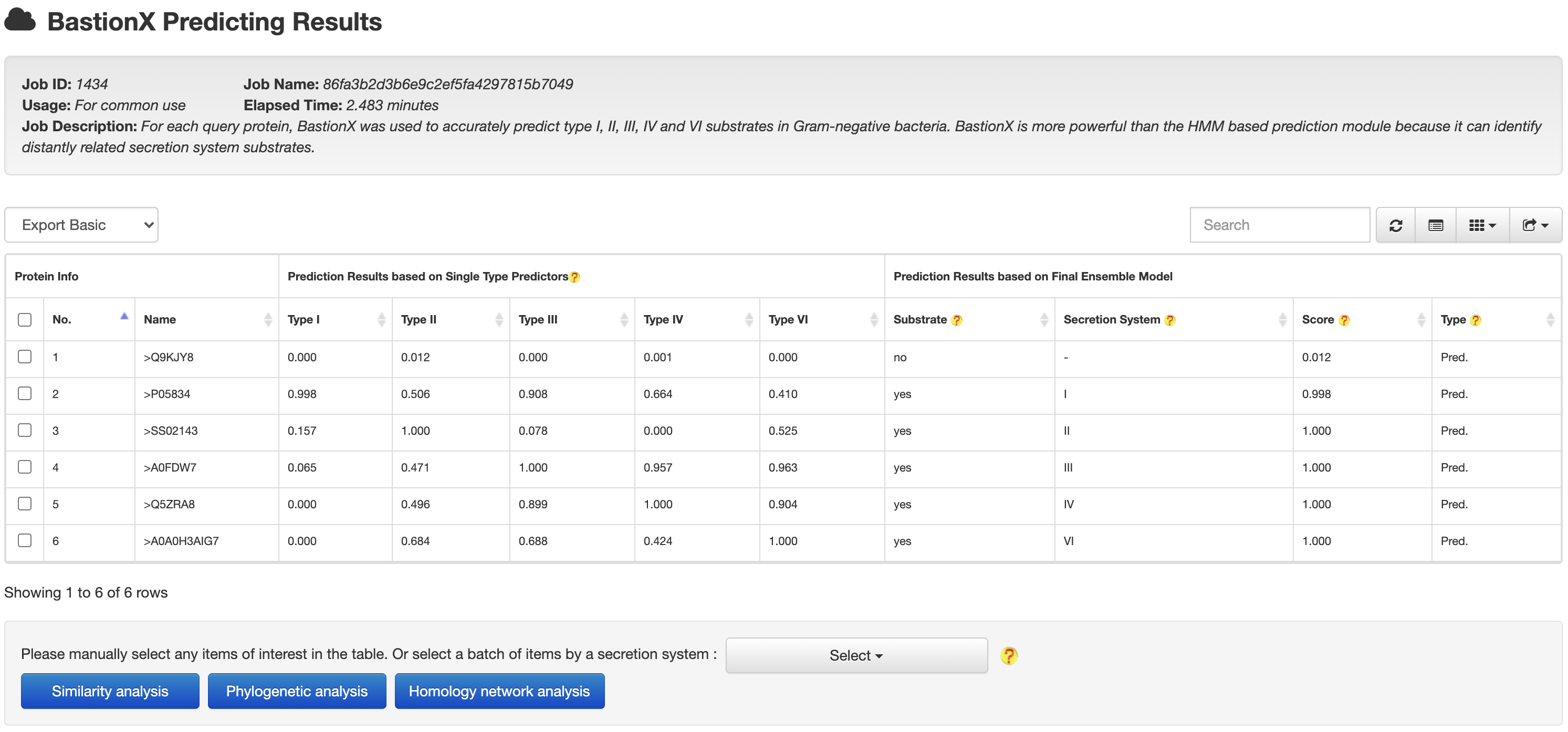
2. PREDICTION
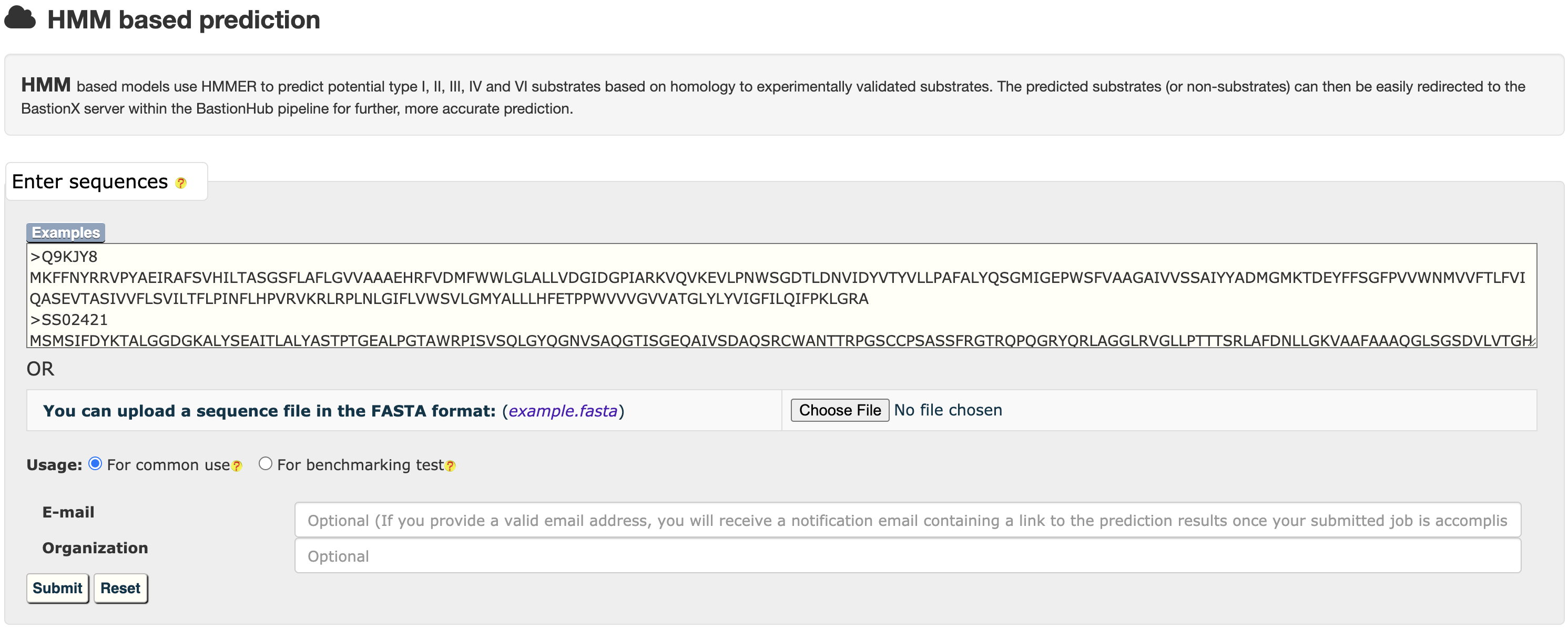
2.1 HMM based prediction
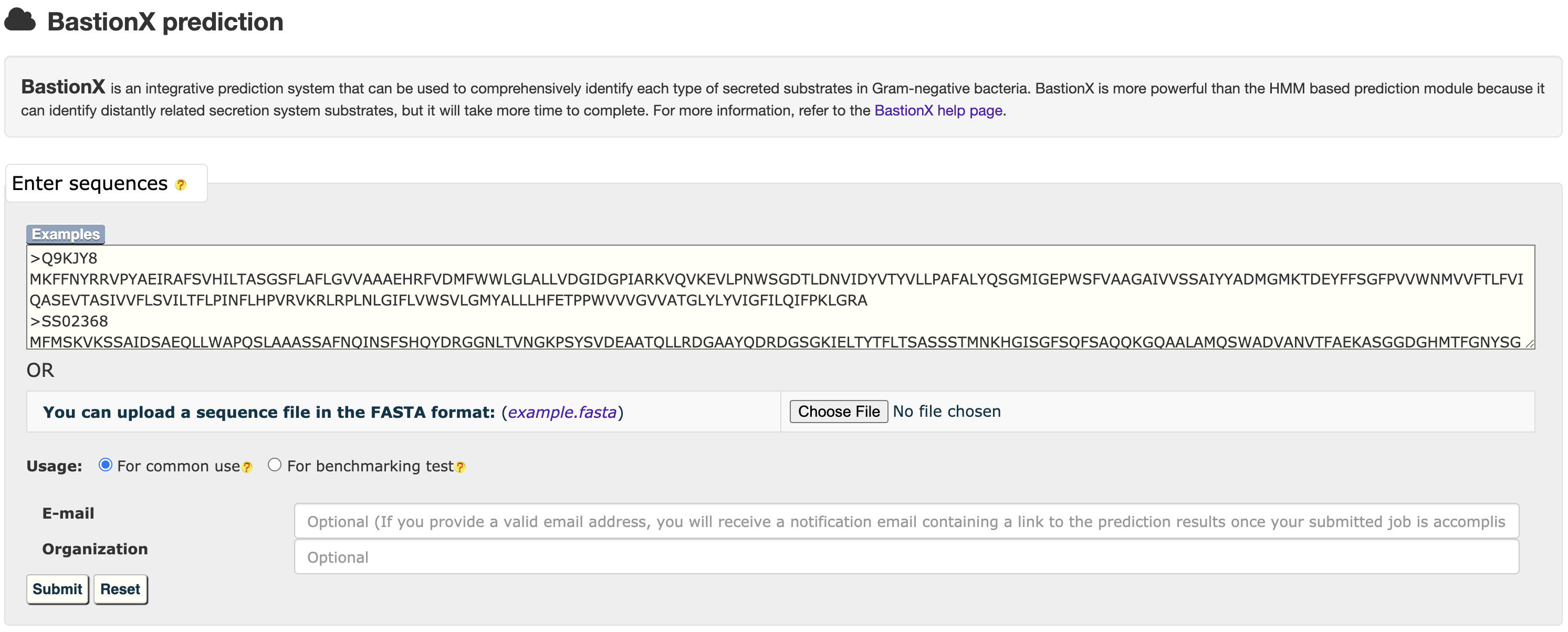
2.2 BastionX
3. RELATIONSHIP ANALYSIS
BastionHub provides three modules for users to analyze relationships between predicted and known substrates, including similarity analysis, phylogenetic analysis and homology network anlysis.
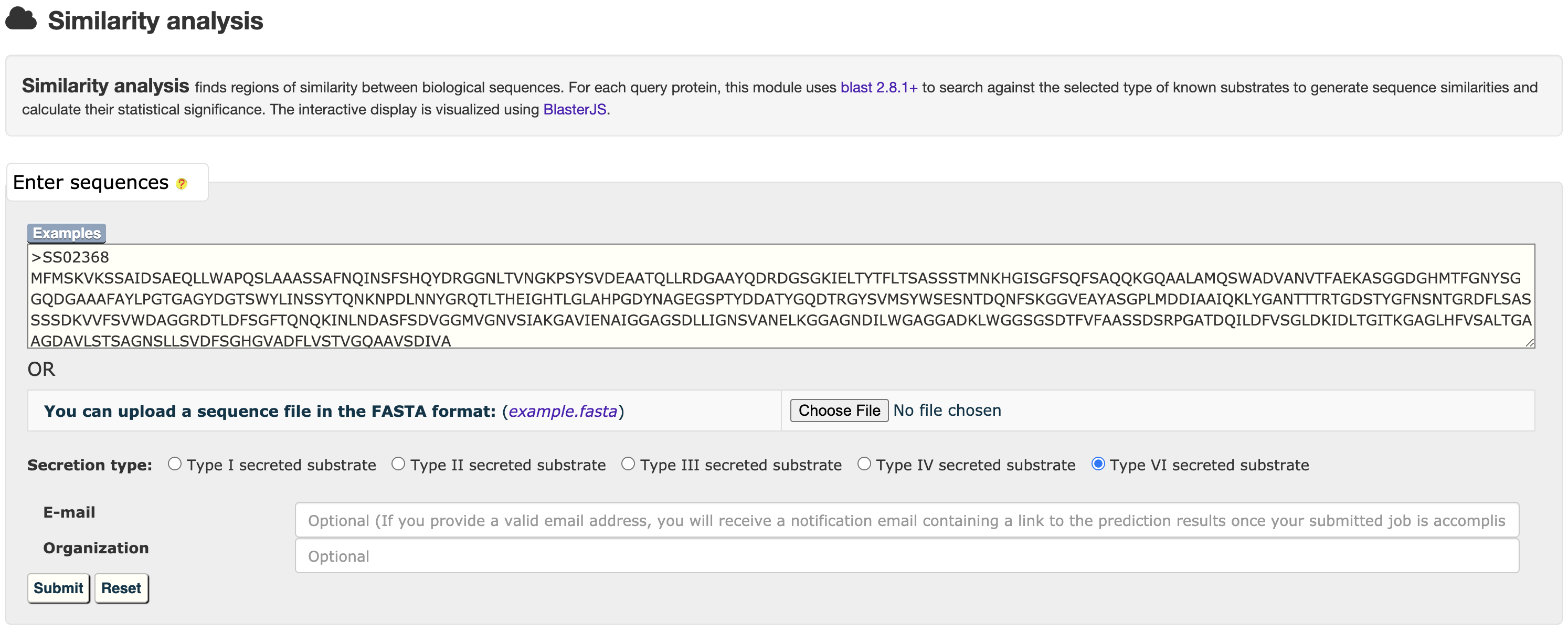
3.1 Similarity analysis
3.2 Phylogenetic analysis
3.3 Homology network analysis
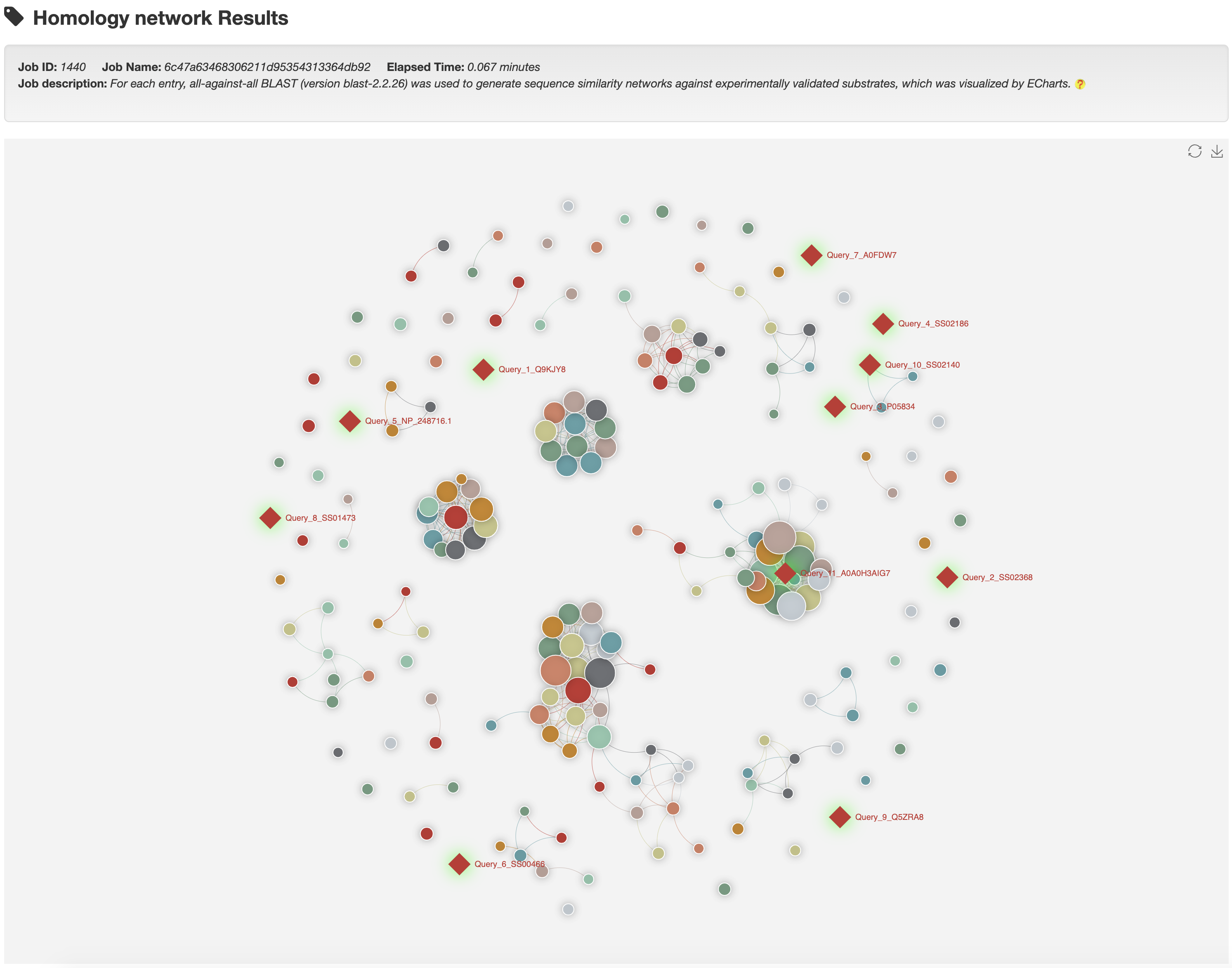
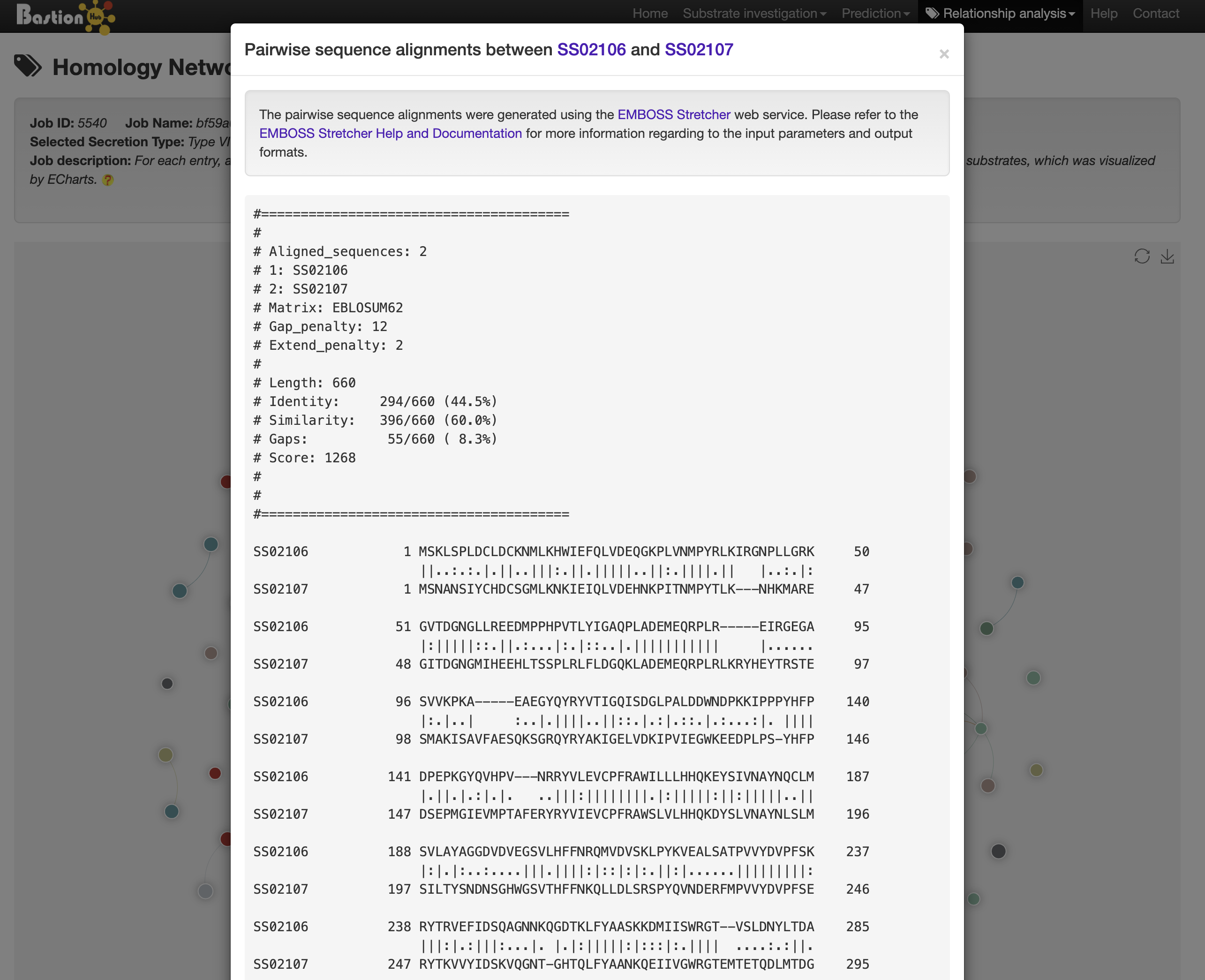
Clicking any edge in the network will show the pairwise sequence alignments between the two linked known substrates.
4. ANALYSIS PIPELINES
BastionHub provides options for users to transfer between different modules, including from prediction to prediction modules, from prediction to analysis modules and from any computational modules to detailed pages of homologous known substrates.
4.1 From prediction to prediction
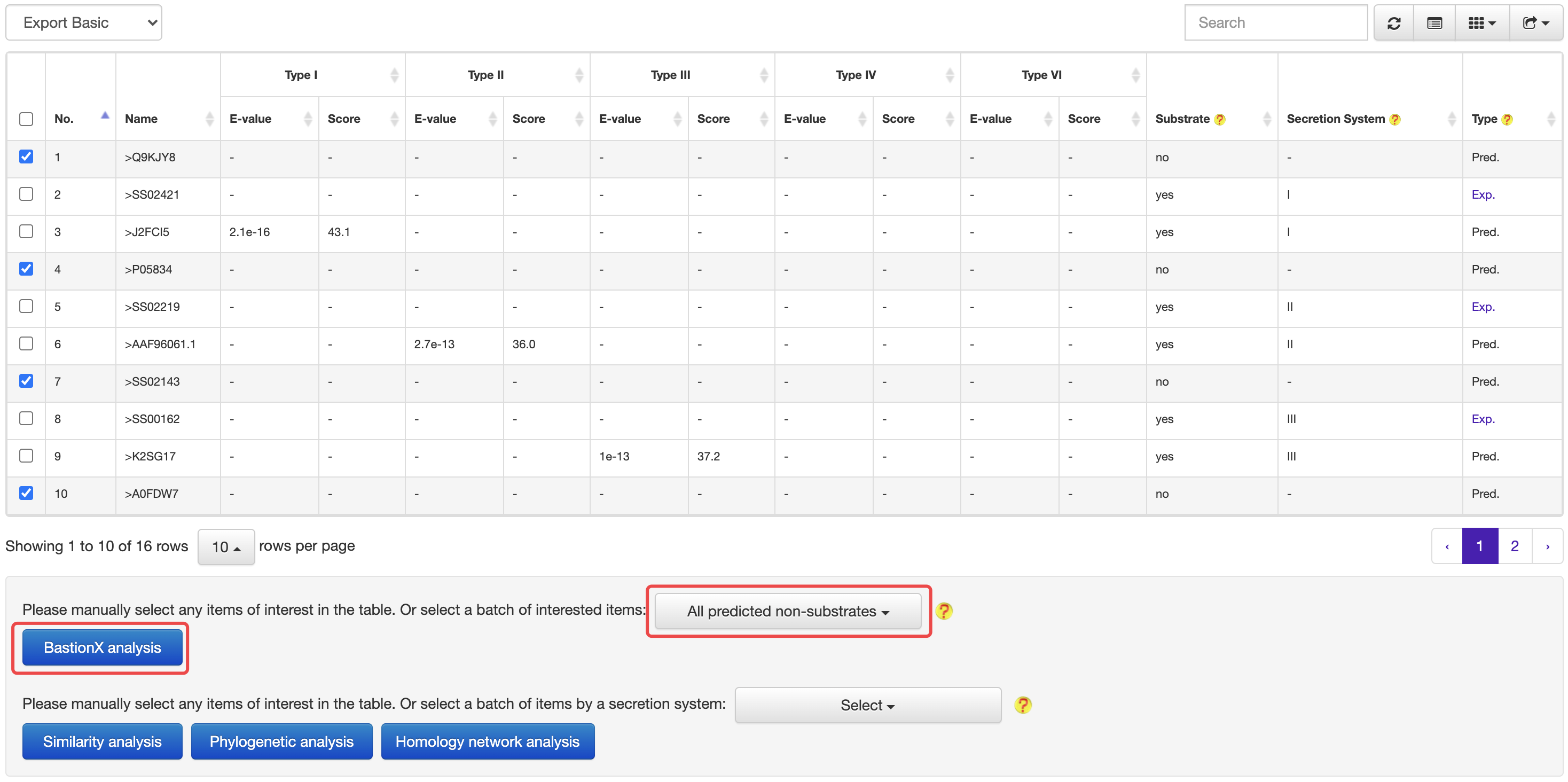
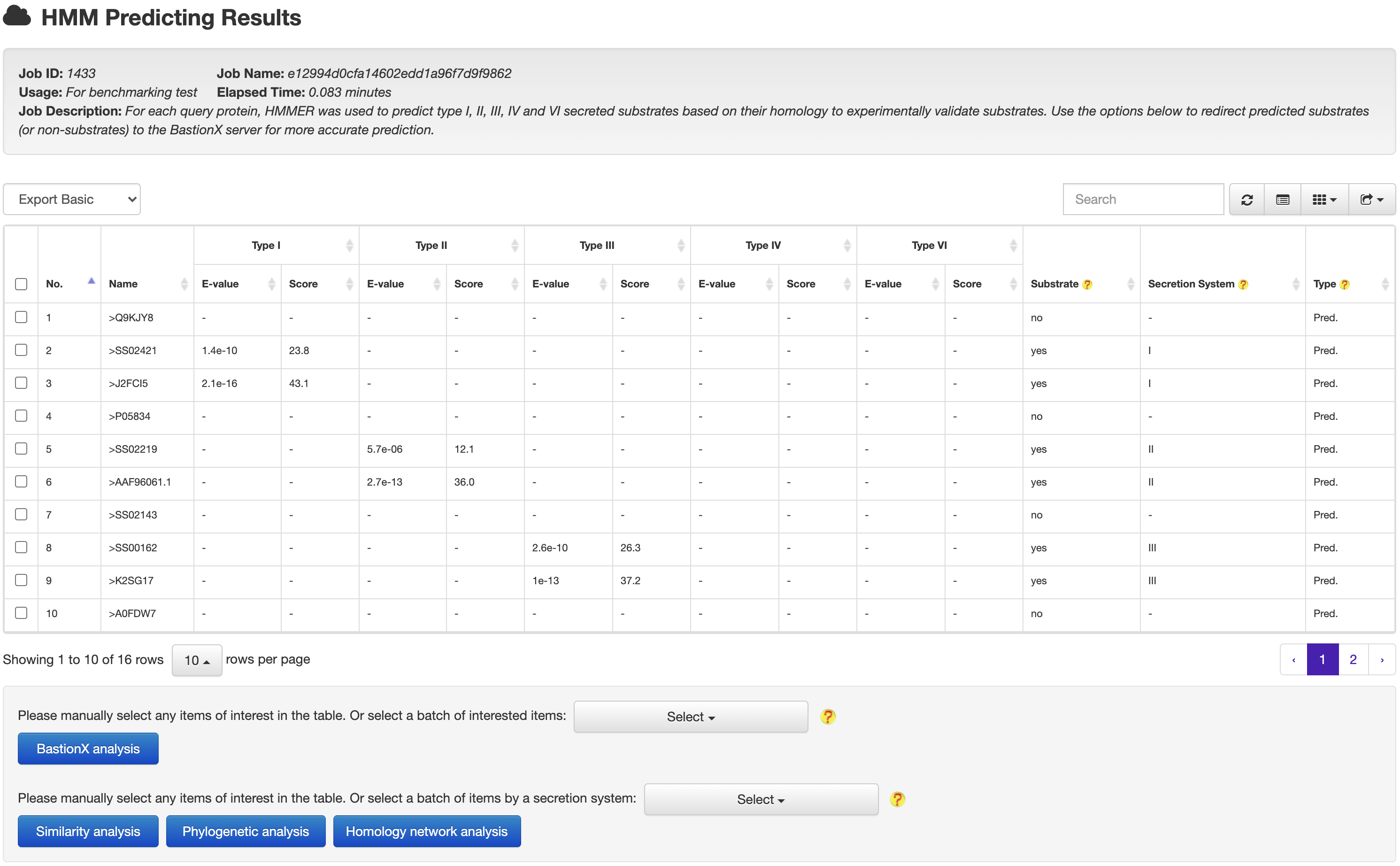
At the HMM based prediction results page, users could easily select a set of target proteins, and redirect them into a BastionX prediction module for more accurate prediction.
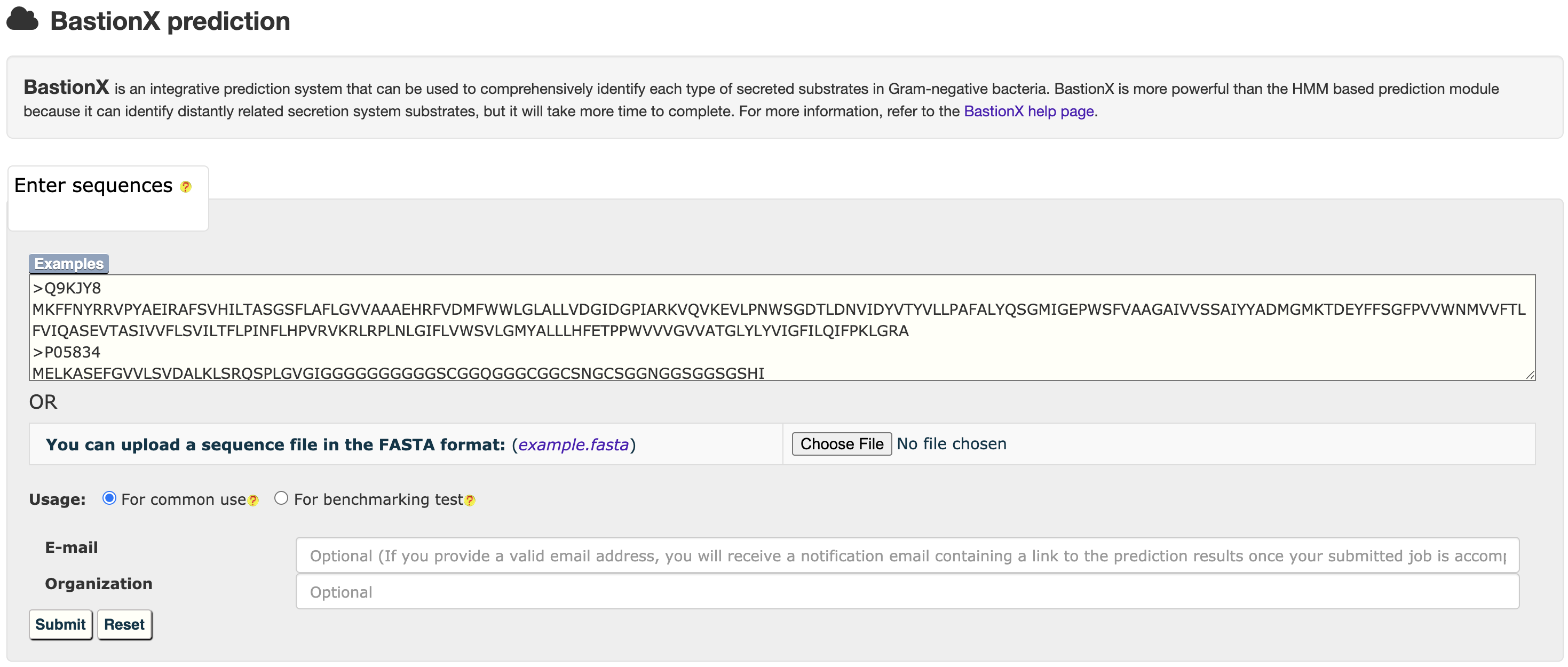
4.2 From prediction to relationship
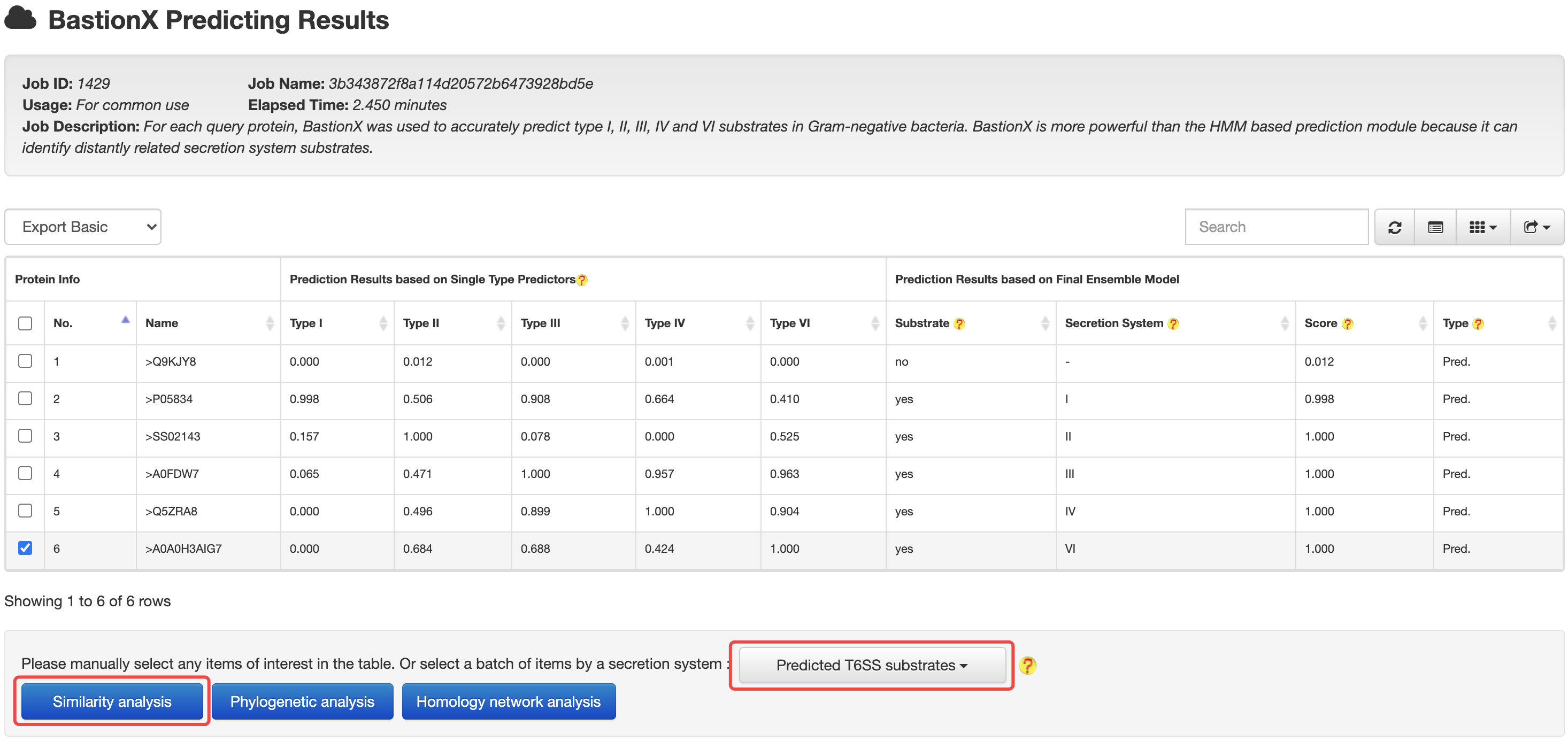
At the HMM or BastionX prediction results page, users could easily select a set of target proteins, and redirect them into a relationship analysis module.
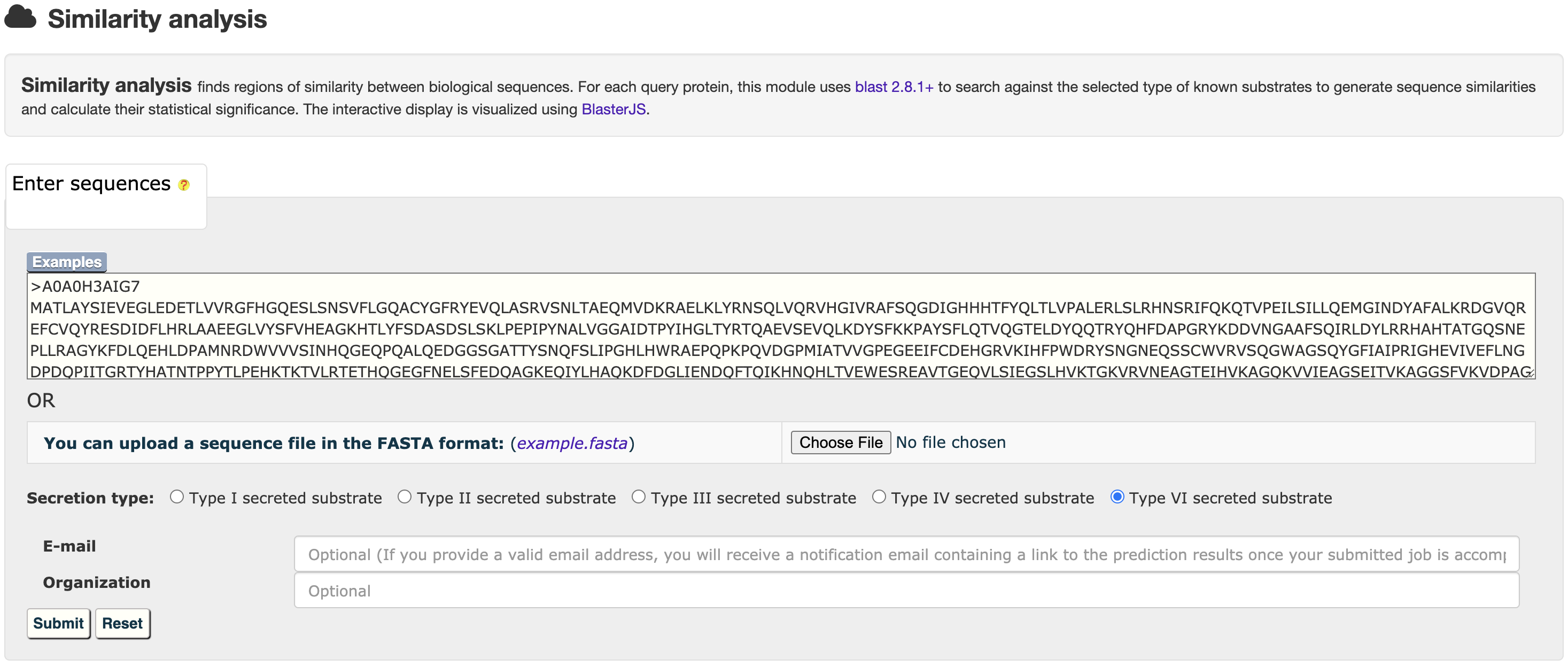
and then get the relationship analysis results:
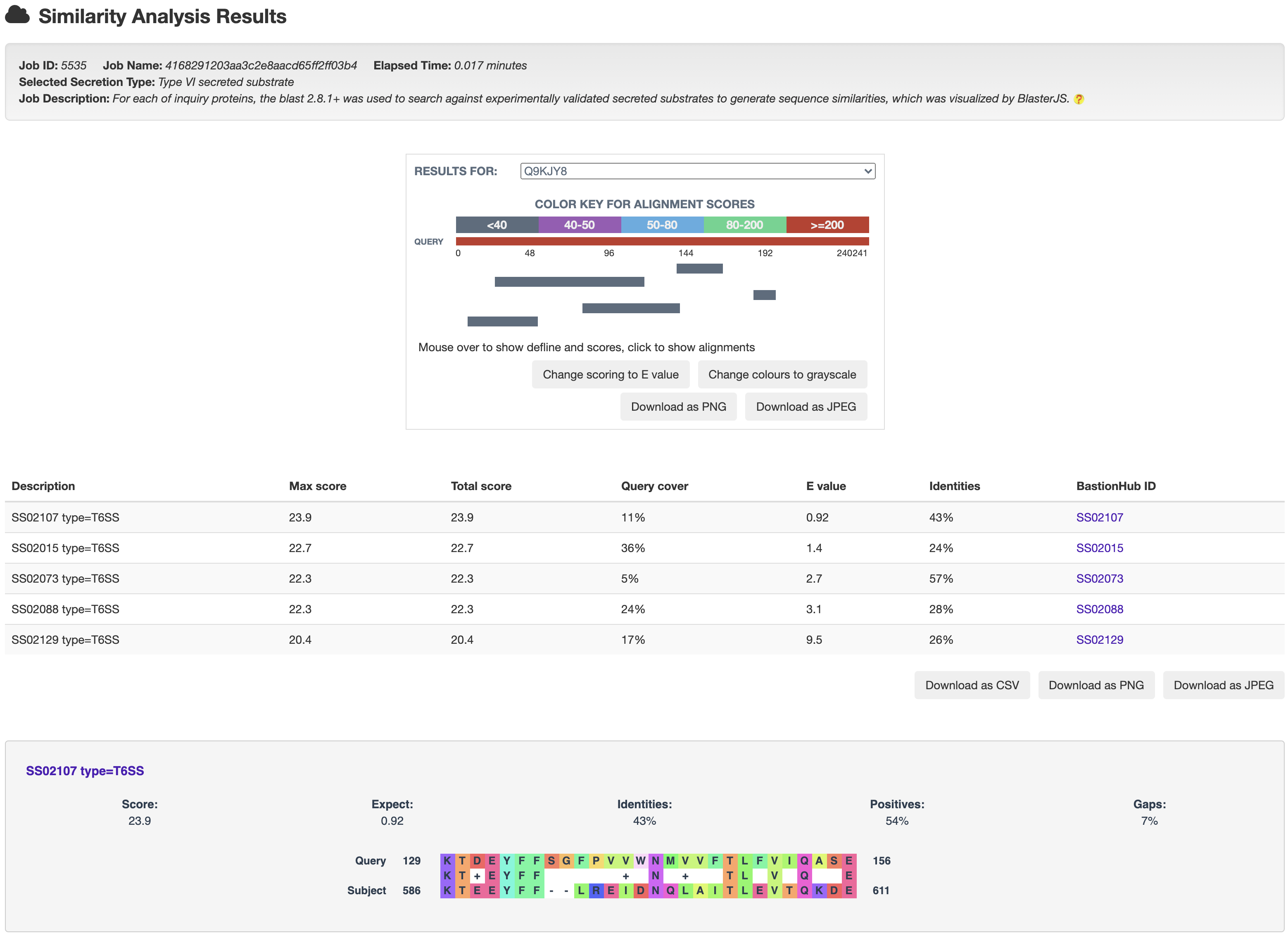
4.3 From results to known substrates
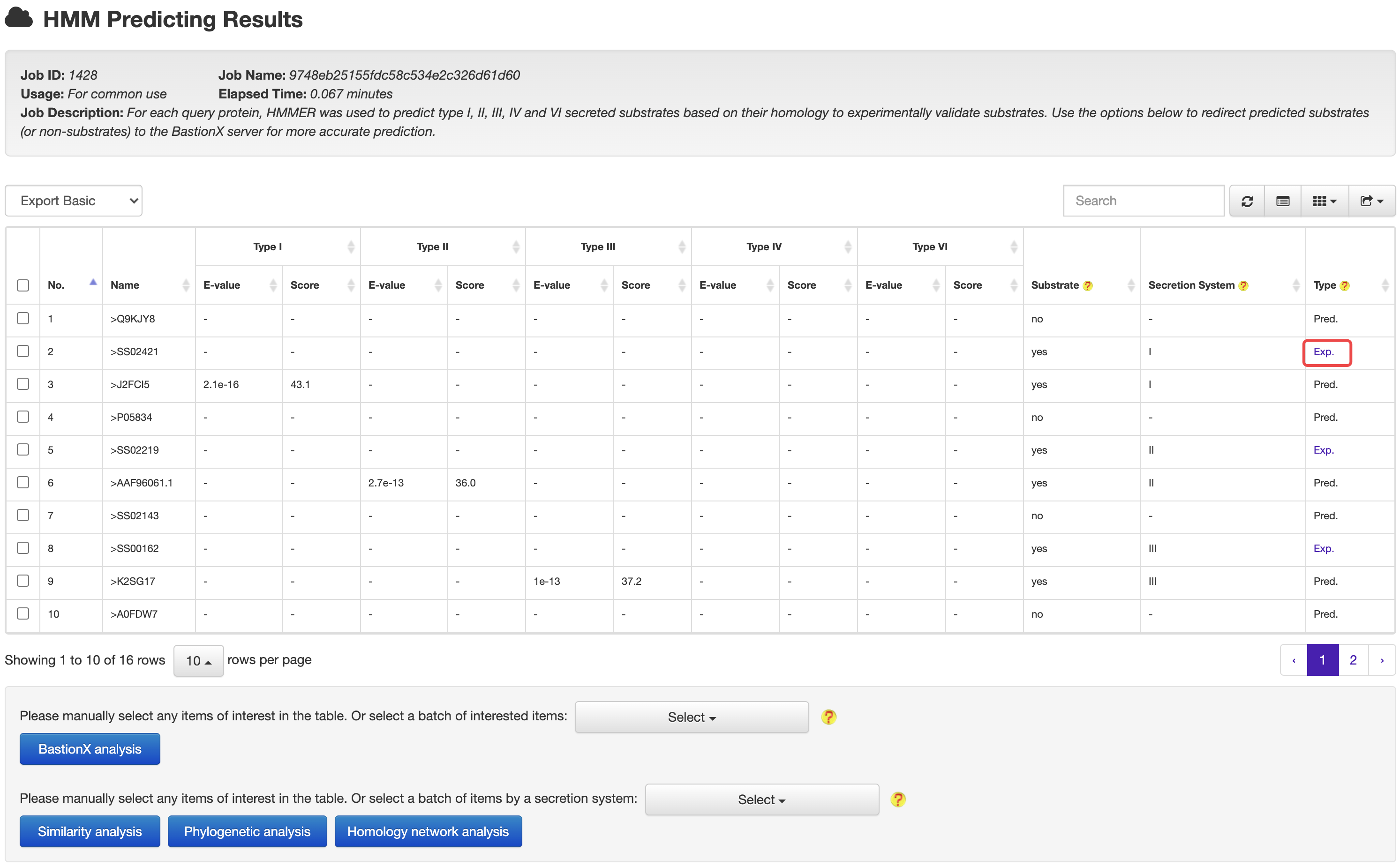
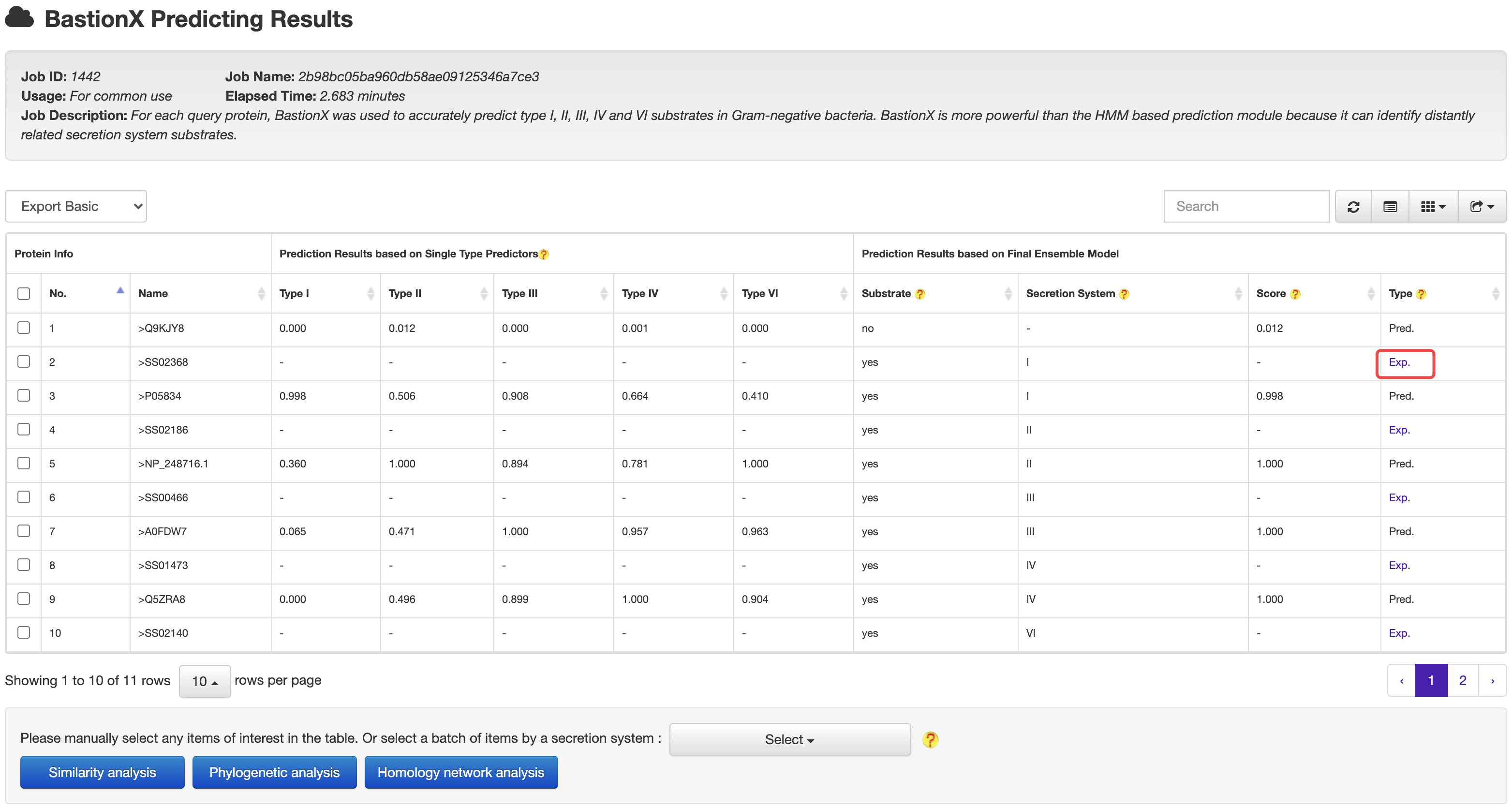
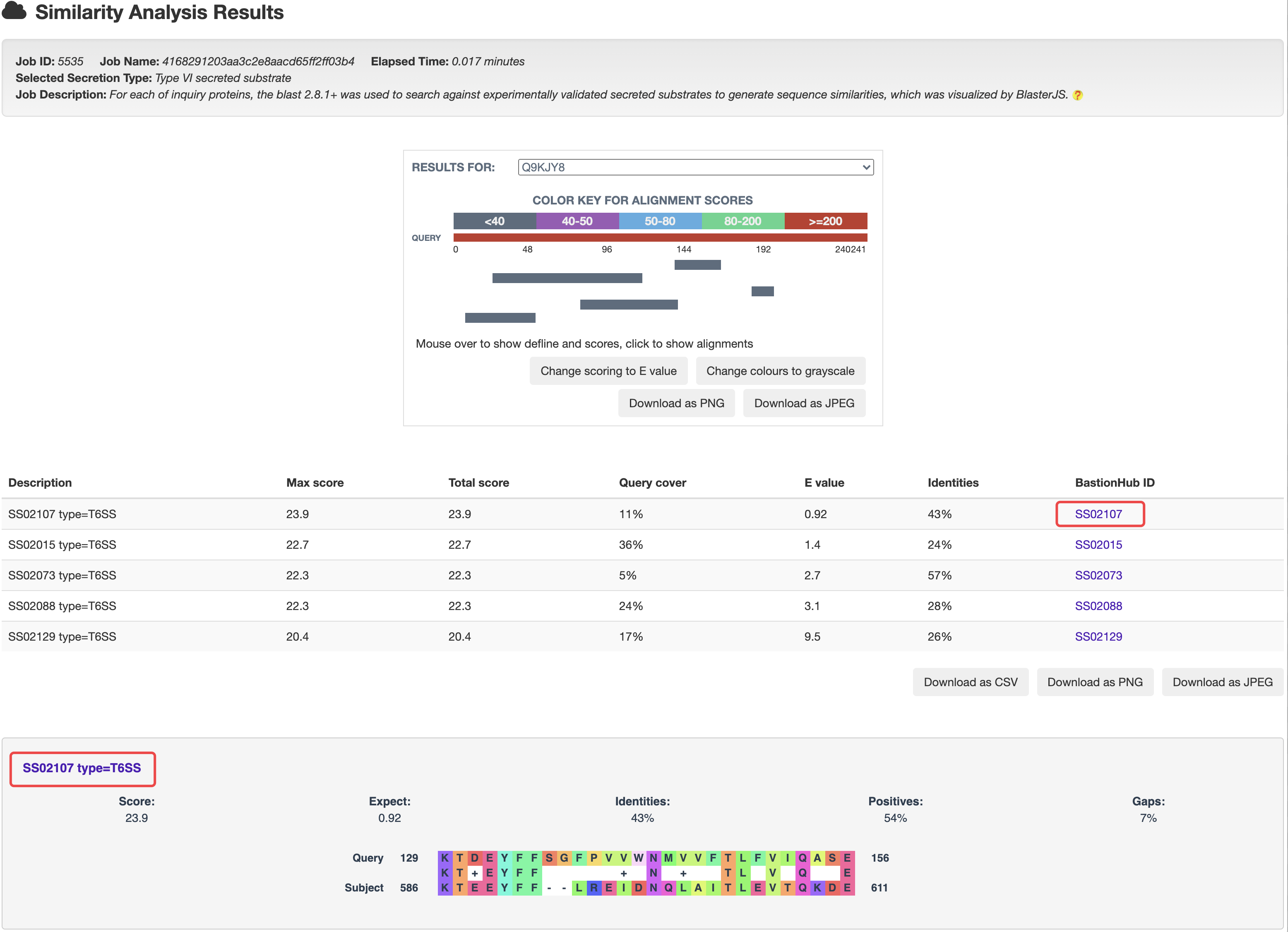
At the prediction or relationship analysis results pages, users could click links to access the detailed information of associated known substrates.